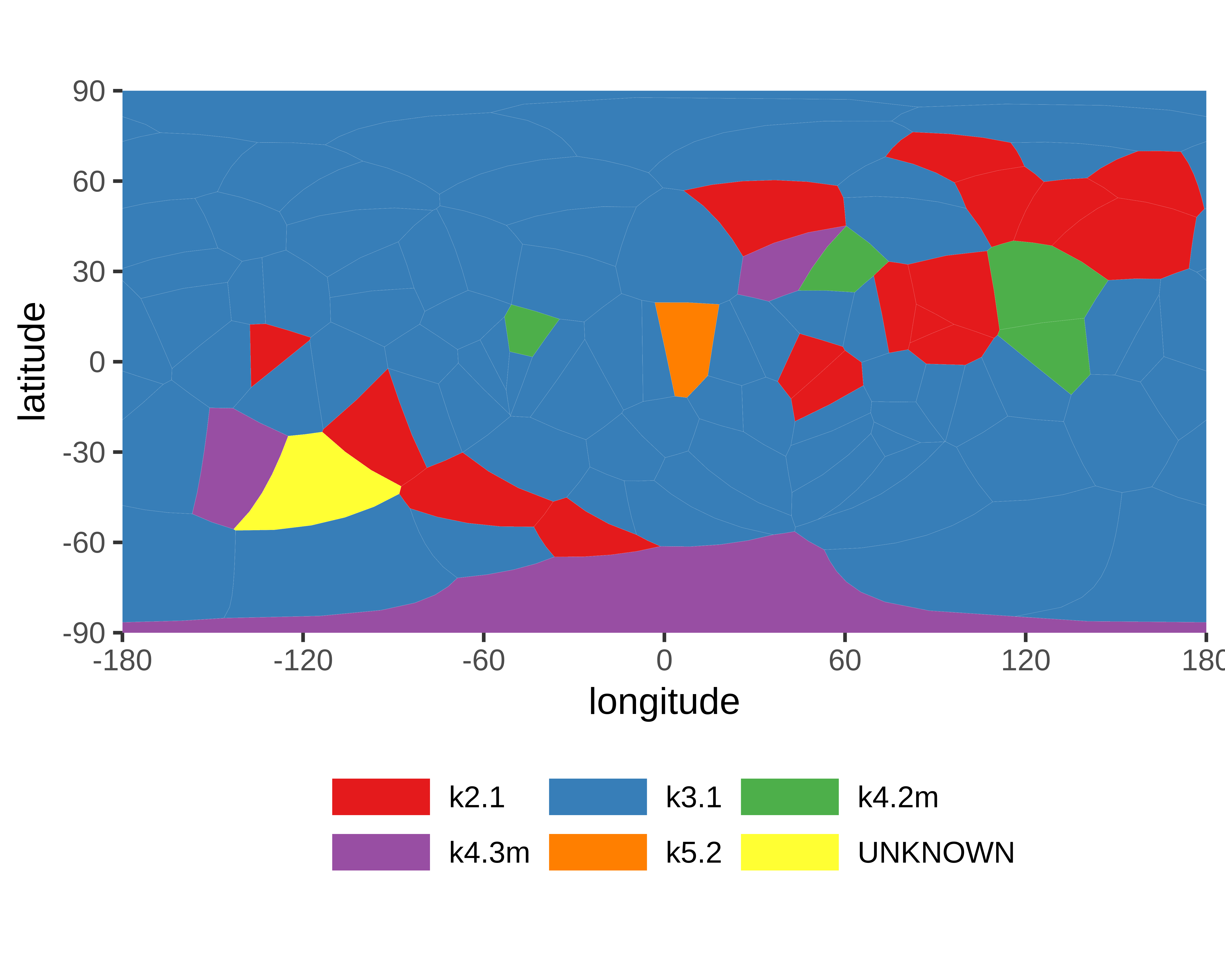
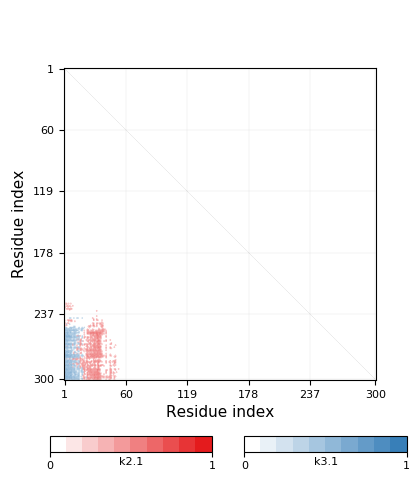
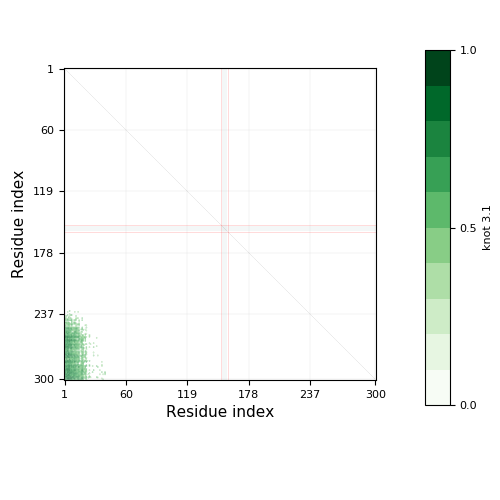
|
|
|
|
K +31
|
Met1 ... Ile118 <-> Bridging ionSr403 <-> Asp127 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Ala47 <-> Bridging ionNa401 <-> Ser51 ... Ile118 <-> Bridging ionSr403 <-> Asp127 ... Glu213 <-> Bridging ionNa401 <-> Thr50 ... Ala47 |
ion-based |
|
|
|
|
|
K +31
|
Glu54 ... Ile118 <-> Bridging ionSr403 <-> Asp127 ... Glu213 <-> Bridging ionNa402 <-> Glu54 |
ion-based |
|
|
|
|
|
K +31
|
Ala47 <-> Bridging ionNa401 <-> Ser51 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Glu213 <-> Bridging ionNa401 <-> Thr50 ... Ala47 |
ion-based |
|
|
|
|
|
K +31
|
Thr50 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Glu213 <-> Bridging ionNa401 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Thr50 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Ser236 <-> Bridging ionNa401 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Ile118 <-> Bridging ionSr403 <-> Asp127 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Ile118 <-> Bridging ionSr403 <-> Asp127 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Thr50 <-> Bridging ionNa401 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Thr50 <-> Bridging ionNa401 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Glu54 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Thr50 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Thr209 <-> Bridging ionNa402 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Thr50 <-> Bridging ionNa402 <-> Glu54 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Ala47 <-> Bridging ionNa401 <-> Ser51 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Glu213 <-> Bridging ionNa401 <-> Thr50 ... Ala47 |
ion-based |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Thr50 <-> Bridging ionNa402 <-> Glu54 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Ala47 <-> Bridging ionNa401 <-> Thr50 ... Ala47 |
ion-based |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Thr50 ... Ile118 <-> Bridging ionSr403 <-> Asp127 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Thr50 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Ser236 <-> Bridging ionNa401 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Thr50 <-> Bridging ionNa402 <-> Glu54 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Ser236 <-> Bridging ionNa401 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Thr50 <-> Bridging ionNa401 <-> Glu213 ... Thr209 <-> Bridging ionNa402 <-> Glu54 ... Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Thr50 <-> Bridging ionNa402 <-> Glu54 ... Ile118 <-> Bridging ionSr403 <-> Asp127 ... Glu213 <-> Bridging ionNa401 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Ile118 <-> Bridging ionSr403 <-> Asp127 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Thr50 <-> Bridging ionNa402 <-> Glu54 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Ala47 <-> Bridging ionNa401 <-> Ser51 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Thr209 <-> Bridging ionNa402 <-> Thr50 ... Ala47 |
ion-based |
|
|
|
|
|
K +31
|
Glu54 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Thr209 <-> Bridging ionNa402 <-> Glu54 |
ion-based |
|
|
|
|
|
K +31
|
Thr50 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Thr209 <-> Bridging ionNa402 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Ser51 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Glu54 ... Ile118 <-> Bridging ionSr403 <-> Asp121 ... Glu213 <-> Bridging ionNa402 <-> Glu54 |
ion-based |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Thr50 ... Thr209 <-> Bridging ionNa402 <-> Glu213 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... Ala47 <-> Bridging ionNa401 <-> Thr50 ... Glu54 <-> Bridging ionNa402 <-> Thr209 ... Chain closureArg300 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Thr50 <-> Bridging ionNa401 <-> Glu213 ... Thr209 <-> Bridging ionNa402 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Thr50 <-> Bridging ionNa402 <-> Glu54 ... Glu213 <-> Bridging ionNa401 <-> Thr50 |
ion-based |
|
|
|
|
|
K +31
|
Thr50 <-> Bridging ionNa402 <-> Glu54 ... Asp121 <-> Bridging ionSr403 <-> Asp127 ... Glu213 <-> Bridging ionNa401 <-> Thr50 |
ion-based |
|

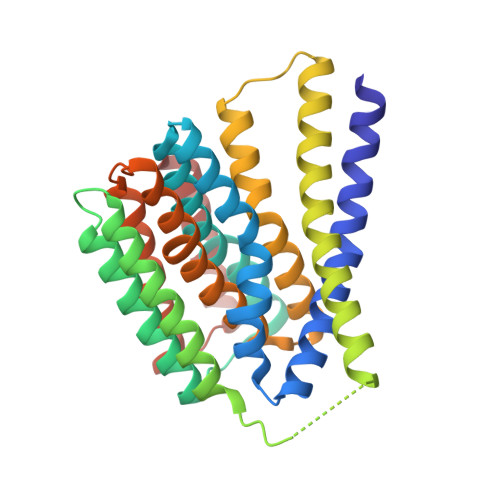
 Genus: 126
Genus: 126