The protein topology database KnotProt collects information about protein structures with open polypeptide chains forming knots or slipknots. The knotting complexity of the catalogued proteins is presented in the form of a matrix diagram (introduced in this pnas paper) that shows users the knot type of the entire polypeptide chain and of each of its subchains. The pattern visible in the matrix gives the knotting fingerprint of a given protein and permits users to determine, for example, the minimal length of the knotted regions (knots’ core size) or the depth of a knot, i.e. how many aminoacids can be removed from either end of the catalogued protein structure before converting it from a knot to a different type of knot. In addition, the database presents extensive information about the biological function of proteins with non-trivial knotting and the families and fold types of these proteins. As an additional feature, the KnotProt database enables users to submit protein or polymer structures and generate their knotting fingerprints.
The data currently deposited in this database is summarized in database statistics.
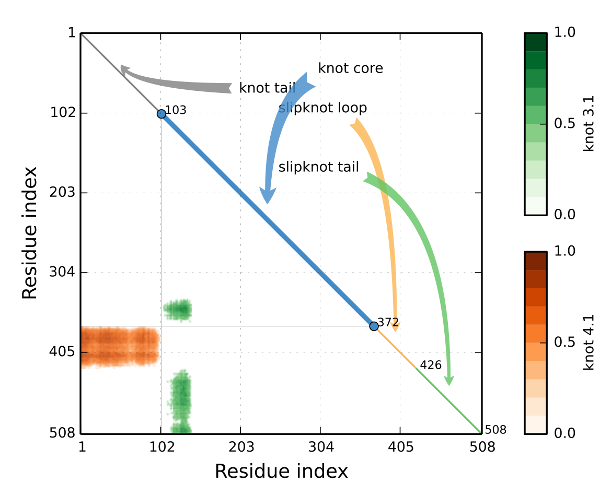
Example of a topological fingerprint with a slipknot motif denoted S31,41,31 detected in a protein.
Number of (slip)knotted chains deposited in the PDB and KnotProt databases.
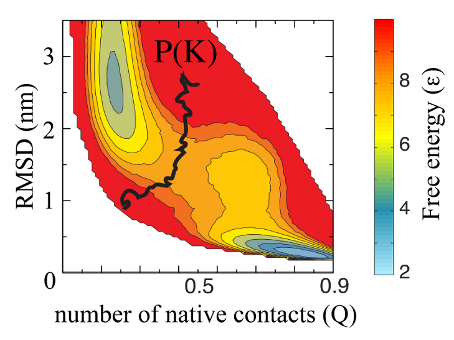
F(Q) Plot
Example of evaluation of data obtained from the webserver, based on time evolution of an artificial knot. The figure shows two-dimensional free energy landscape of knotted proteins, measured by the fraction of native contacts formed Q and RMSD. The black curve shows the contour of knot probability obtained from time dependent protein coordinates (based on results in
this pnas paper).
The KnotProt can be used also as a server to analyze time evolution of knots and slipknots (uploaded as entire trajectories). Moreover, it can detect complexity in open chains which are not deposited in the rcsb pdb.
Authors
This database has been created in a joint collaboration between: Michał Jamróz (University of Warsaw, Faculty of Chemistry), Wanda Niemyska (University of Silesia, Institute of Mathematics), Eric Rawdon (University of St. Thomas, Saint Paul, Department of Mathematics), Andrzej Stasiak (University of Lausanne, Center for Integrative Genomics), Ken Millett (University of California, Santa Barbara, Department of Mathematics), Piotr Sułkowski (University of Warsaw, Faculty of Physics), Joanna Sulkowska (University of Warsaw, Faculty of Chemistry and Centre of New Technologies).
In the update of the database (KnotProt 2.0), apart from the original authors Pawel Dabrowski-Tumanski (University of Warsaw, Faculty of Chemistry, Centre of New Technologies), Pawel Rubach (Warsaw School of Economics,), Dimoklis Goundaroulis (University of Lausanne, Center for Integrative Genomics and SIB Swiss Institute of Bioinformatics), and Julien Dorier (University of Lausanne, Center for Integrative Genomics and Vital-IT, SIB Swiss Institute of Bioinformatics) took part in creation of the database.
Funding
The research leading to creation of this database has been supported by: National Science Center [grant agreement Sonata BIS 2012/07/E/NZ1/01900 to J.S., PRELUDIUM #2016/21/N/NZ1/02848 to P.D.-T.]; Foundation for Polish Science [Homing Plus to J.S., SKILLS/Inter grant agreement 177/UD/SKILLS/2012 co-financed by European Social Fund to P.S., TEAM grant agreement TEAM/2011-7/6, co-financed by the EU European Regional Development Fund operated within the Innovative Economy Operational Program to M.J.]; Ministry of Science and Higher Education [Iuventus Plus programme to P.S.]; National Science Foundation [Division of Mathematical Sciences, #1115722,#1418869 to E.R.]; Swiss National Foundation [31003A_138267 to A.S.], The Leverhulme Trust [RP2013-K-017 to A.S.], and ERC Starting Grant [#335739 to P.S.]. Funding for open access charge: Swiss National Foundation [31003A_138267].


