
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 16-316 | 301 | 1-15 | 471-526 | 317-470 | 15 | 56 | slipknot |
Chain Sequence |
AARSIAATPPKLIVAISVDQFSADLFSEYRQYYTGGLKRLTSEGAVFPRGYQSHAATETCPGHSTILTGSRPSRTGIIANNWFDLDAKREDKNLYCAEDESQPGSSSDKYEASPLHLKVPTLGGRMKAANPATRVVSVAGKDRAAIMMGGATADQVWWLGGPQGYVSYKGVAPTPLVTQVNQAFAQRLAQPNPGFELPAQCVSKDFPVQAGNRTVGTGRFARDAGDYKGFRISPEQDAMTLAFAAAAIENMQLGKQAQTDIISIGLSATDYVGHTFGTEGTESCIQVDRLDTELGAFFDKLDKDGIDYVVVLTADHGGHDLPERHRMNAMPMEQRVDMALTPKALNATIAEKAGLPGKKVIWSDGPSGDIYYDKGLTAAQRARVETEALKYLRAHPQVQTVFTKAEIAATPSPSGPPESWSLIQEARASFYPSRSGDLLLLLKPRVMSIPEQAVMGSVATHGSPWDTDRRVPILFWRKGMQHFEQPLGVETVDILPSLAALIKLPVPKDQIDGRCLDLVAGKDDSC
|
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
- 31 | 20-119 | 100 | 1-19, 120-174 | 19 | 55 | knot |
| Fingerprint | Knot forming loop | Loop type | ||||
|---|---|---|---|---|---|---|
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... His304 <-> Bridging ionZn1561 <-> His491 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... His304 <-> Bridging ionZn1561 <-> His491 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 <-> Bridging ionCys556 <-> Cys545 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 <-> Bridging ionCys556 <-> Cys545 ... His491 <-> Bridging ionZn1561 <-> His304 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 <-> Bridging ionCys556 <-> Cys545 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 <-> Bridging ionCys556 <-> Cys545 ... His491 <-> Bridging ionZn1561 <-> His304 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Cys314 <-> Cys231 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn1561 <-> His491 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... His304 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Cys314 <-> Cys231 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn1561 <-> His491 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... His304 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Gly551 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... His304 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Gly551 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Gly551 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... His304 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Gly551 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Val549 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Val549 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Val549 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Val549 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Gly551 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... His304 <-> Bridging ionZn1561 <-> His491 ... Gly551 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Gly551 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... His304 <-> Bridging ionZn1561 <-> His491 ... Gly551 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Asp553 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Asp553 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Asp553 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Asp553 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Gly551 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Gly551 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Gly551 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Gly551 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 <-> Bridging ionCys556 <-> Cys545 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 <-> Bridging ionCys556 <-> Cys545 ... His491 <-> Bridging ionZn1561 <-> His304 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 <-> Bridging ionCys556 <-> Cys545 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 <-> Bridging ionCys556 <-> Cys545 ... His491 <-> Bridging ionZn1561 <-> His304 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Cys314 <-> Cys231 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Cys314 <-> Cys231 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Cys314 <-> Cys231 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Gly551 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Gly551 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Cys314 <-> Cys231 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Gly551 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn1561 <-> His491 ... Val549 <-> Bridging ionCa566 <-> Gly551 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Val549 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Val549 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Val549 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Val549 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Cys314 <-> Cys231 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Gly551 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> His346 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn1561 <-> His491 ... Gly551 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Cys314 <-> Cys231 ... Asp300 <-> Bridging ionZn1561 <-> His491 ... Gly551 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Ala31 ... Thr89 <-> Bridging ionZn1559 <-> Asp345 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn1561 <-> His491 ... Gly551 <-> Bridging ionCa566 <-> Asp553 ... Chain closureCys556 <-> Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Asp553 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Asp553 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Asp553 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Asp553 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Gly551 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Gly551 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Gly551 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic | |||
|
|
K -31 | Chain closureAla31 <-> Cys556 ... Gly551 <-> Bridging ionCa566 <-> Asp547 ... His491 <-> Bridging ionZn1561 <-> His304 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn1559 <-> Thr89 ... Ala31 |
probabilistic |
Chain Sequence |
AARSIAATPPKLIVAISVDQFSADLFSEYRQYYTGGLKRLTSEGAVFPRGYQSHAATETCPGHSTILTGSRPSRTGIIANNWFDLDAKREDKNLYCAEDESQPGSSSDKYEASPLHLKVPTLGGRMKAANPATRVVSVAGKDRAAIMMGGATADQVWWLGGPQGYVSYKGVAPTPLVTQVNQAFAQRLAQPNPGFELPAQCVSKDFPVQAGNRTVGTGRFARDAGDYKGFRISPEQDAMTLAFAAAAIENMQLGKQAQTDIISIGLSATDYVGHTFGTEGTESCIQVDRLDTELGAFFDKLDKDGIDYVVVLTADHGGHDLPERHRMNAMPMEQRVDMALTPKALNATIAEKAGLPGKKVIWSDGPSGDIYYDKGLTAAQRARVETEALKYLRAHPQVQTVFTKAEIAATPSPSGPPESWSLIQEARASFYPSRSGDLLLLLKPRVMSIPEQAVMGSVATHGSPWDTDRRVPILFWRKGMQHFEQPLGVETVDILPSLAALIKLPVPKDQIDGRCLDLVAGKDDSC
|
Knotoid cutoff: 0.5
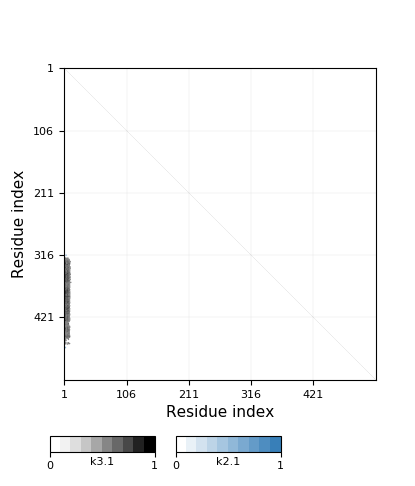
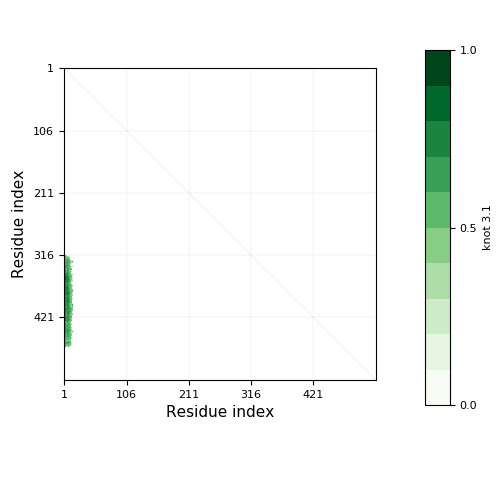
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
3.1 | 17-317 | 301 | 1-16 | 471-526 | 318-470 | 16 | 56 | slipknot | ||
| view details |
|
2.1 | 9-470 | 462 | 1-8 | 473-526 | 471-472 | 8 | 54 | slipknot | ||
| view details |
|
2.1 | 18-404 | 387 | 1-16, 426-526 | 17-17, 405-425 | 16 | 101 | slipknot |
| sequence length |
526
|
| structure length |
526
|
| publication title |
X-ray structure reveals a new class and provides insight into evolution of alkaline phosphatases
pubmed doi rcsb |
| molecule tags |
Hydrolase
|
| molecule keywords |
Alkaline phosphatase
|
| source organism |
Sphingomonas sp.
|
| total genus |
 Genus: 191 Genus: 191
|
| ec nomenclature |
ec
3.1.3.1: Alkaline phosphatase. |
| pdb deposition date | 2010-12-22 |
| KnotProt deposition date | 2018-10-19 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF01663 | Phosphodiest | Type I phosphodiesterase / nucleotide pyrophosphatase |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...