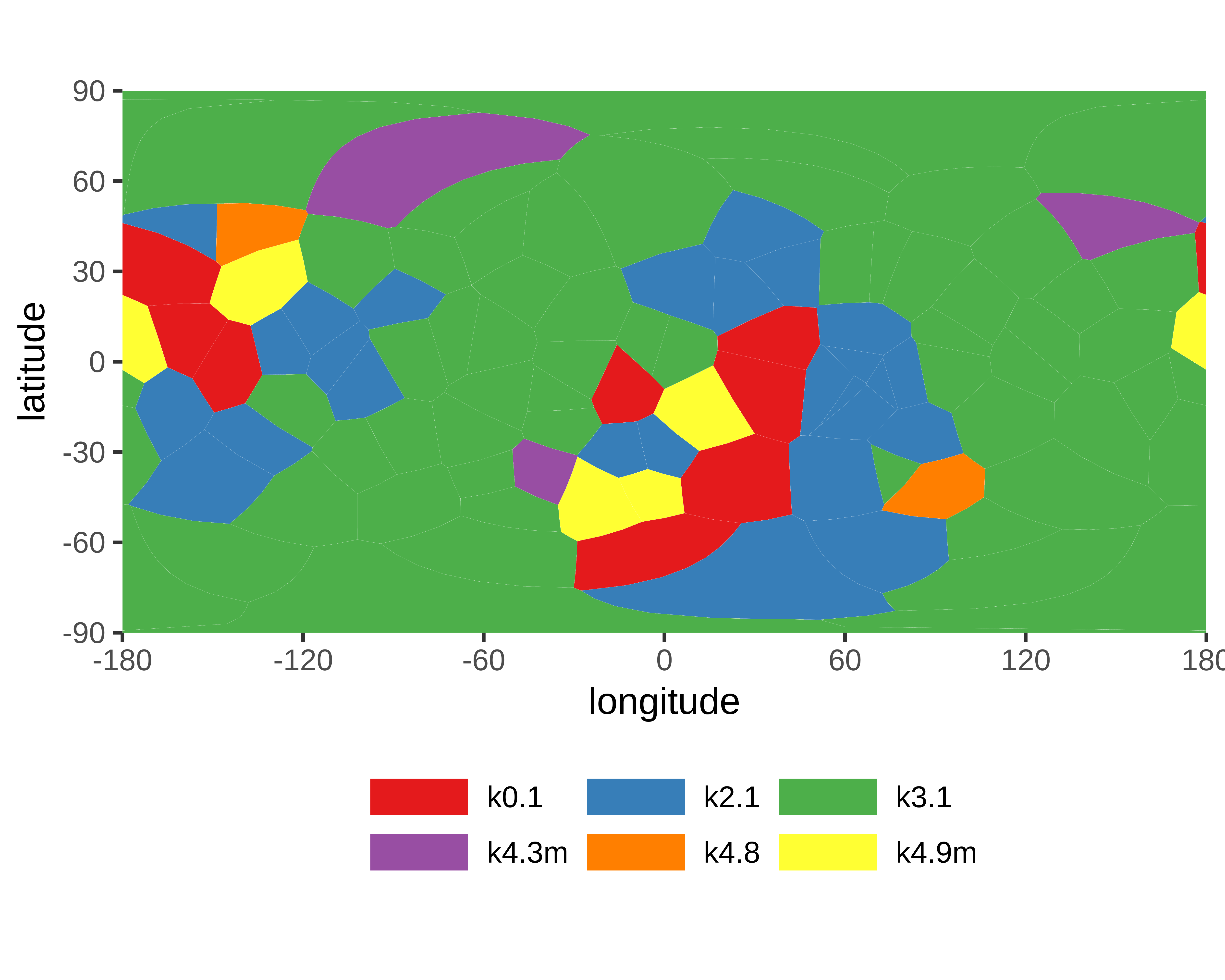
|
|
|
|
K +31
|
Met1 ... Val135 <-> Bridging ionHgb305 <-> Gln137 ... Chain closureLys261 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... His94 <-> Bridging ionZn301 <-> His96 ... Chain closureLys261 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... His96 <-> Bridging ionZn301 <-> His119 ... Chain closureLys261 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... His94 <-> Bridging ionZn301 <-> His119 ... Chain closureLys261 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureMet1 <-> Lys261 ... Cys206 <-> Bridging ionHgb305 <-> Val135 ... Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureMet1 <-> Lys261 ... Cys206 <-> Bridging ionHgb305 <-> Gln137 ... Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... His94 <-> Bridging ionZn301 <-> His96 ... Val135 <-> Bridging ionHgb305 <-> Gln137 ... Chain closureLys261 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... His96 <-> Bridging ionZn301 <-> His119 ... Val135 <-> Bridging ionHgb305 <-> Gln137 ... Chain closureLys261 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Met1 ... His94 <-> Bridging ionZn301 <-> His119 ... Val135 <-> Bridging ionHgb305 <-> Gln137 ... Chain closureLys261 <-> Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureMet1 <-> Lys261 ... Cys206 <-> Bridging ionHgb305 <-> Val135 ... His96 <-> Bridging ionZn301 <-> His94 ... Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureMet1 <-> Lys261 ... Cys206 <-> Bridging ionHgb305 <-> Val135 ... His119 <-> Bridging ionZn301 <-> His96 ... Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureMet1 <-> Lys261 ... Cys206 <-> Bridging ionHgb305 <-> Val135 ... His119 <-> Bridging ionZn301 <-> His94 ... Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureMet1 <-> Lys261 ... Cys206 <-> Bridging ionHgb305 <-> Gln137 ... His96 <-> Bridging ionZn301 <-> His94 ... Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureMet1 <-> Lys261 ... Cys206 <-> Bridging ionHgb305 <-> Gln137 ... His119 <-> Bridging ionZn301 <-> His96 ... Met1 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureMet1 <-> Lys261 ... Cys206 <-> Bridging ionHgb305 <-> Gln137 ... His119 <-> Bridging ionZn301 <-> His94 ... Met1 |
probabilistic |
|

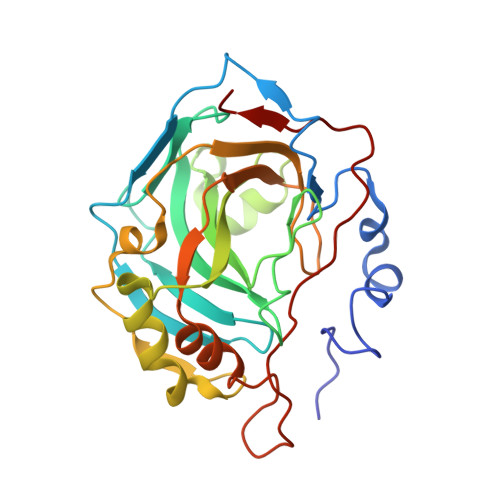
 Genus: 79
Genus: 79