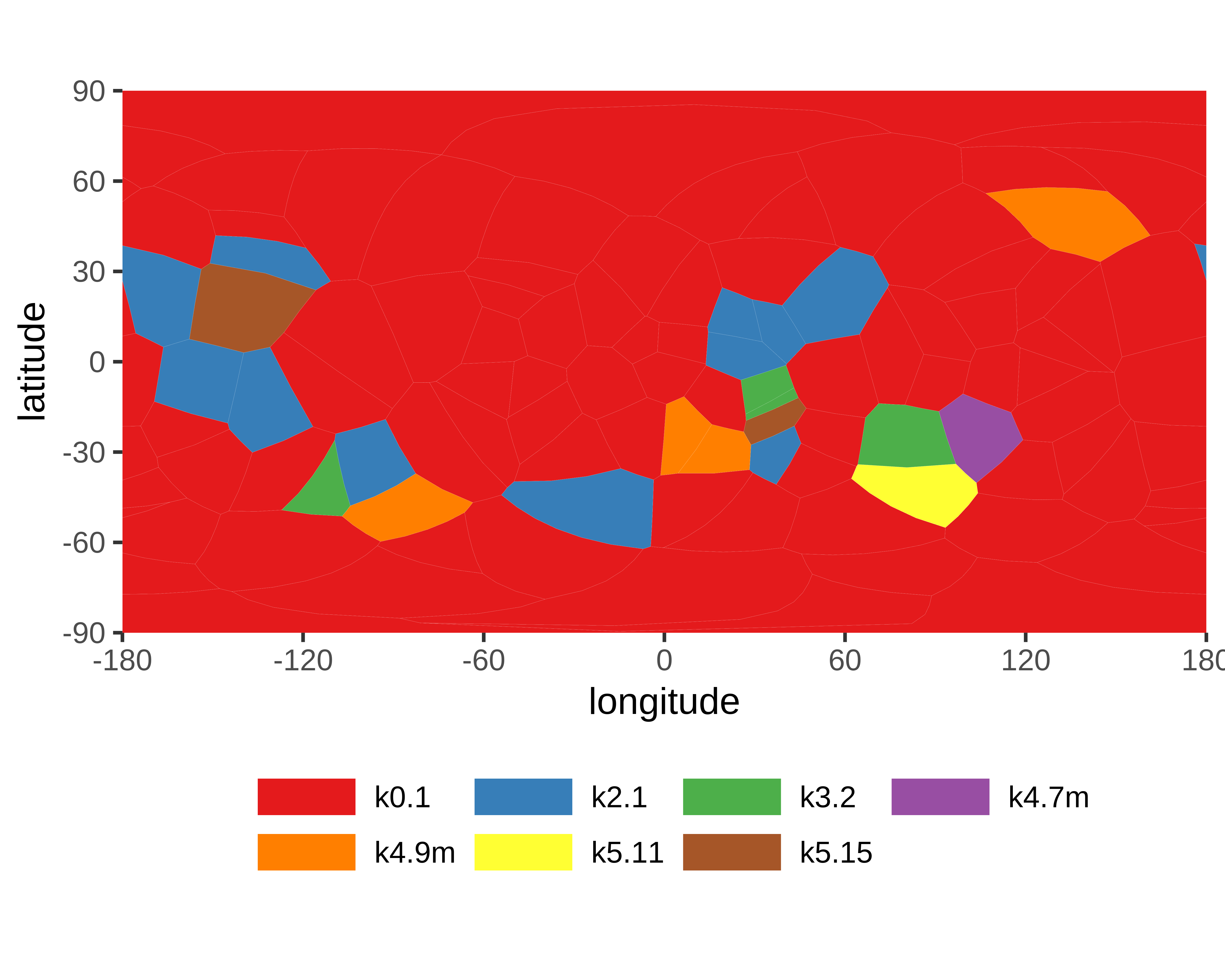
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Ala53 <-> Bridging ionMg603 <-> Leu51 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Ala53 <-> Bridging ionMg603 <-> Ile50 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Ala53 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Leu51 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Ile50 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Ala53 <-> Bridging ionMg603 <-> Leu51 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Ala53 <-> Bridging ionMg603 <-> Ile50 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Ala53 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Leu51 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... His468 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Ile50 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Ala53 <-> Bridging ionMg603 <-> Leu51 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Ala53 <-> Bridging ionMg603 <-> Ile50 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Ala53 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Leu51 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> Asp426 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Ile50 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Ala53 <-> Bridging ionMg603 <-> Leu51 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Ala53 <-> Bridging ionMg603 <-> Ile50 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Ala53 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Leu51 ... Ala20 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla20 <-> His549 ... His485 <-> Bridging ionMn604 <-> His430 ... Asp467 <-> Bridging ionZn605 <-> Ser86 ... Thr55 <-> Bridging ionMg603 <-> Ile50 ... Ala20 |
probabilistic |
|

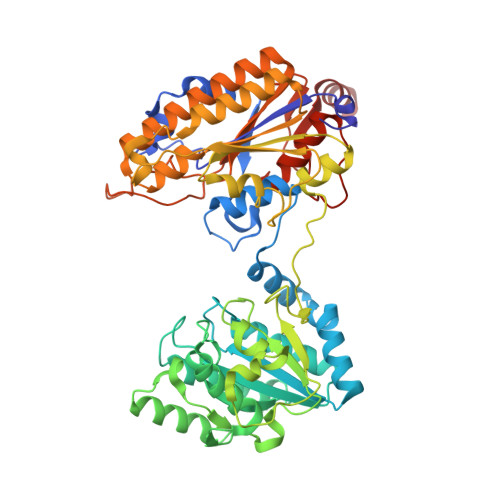
 Genus: 195
Genus: 195