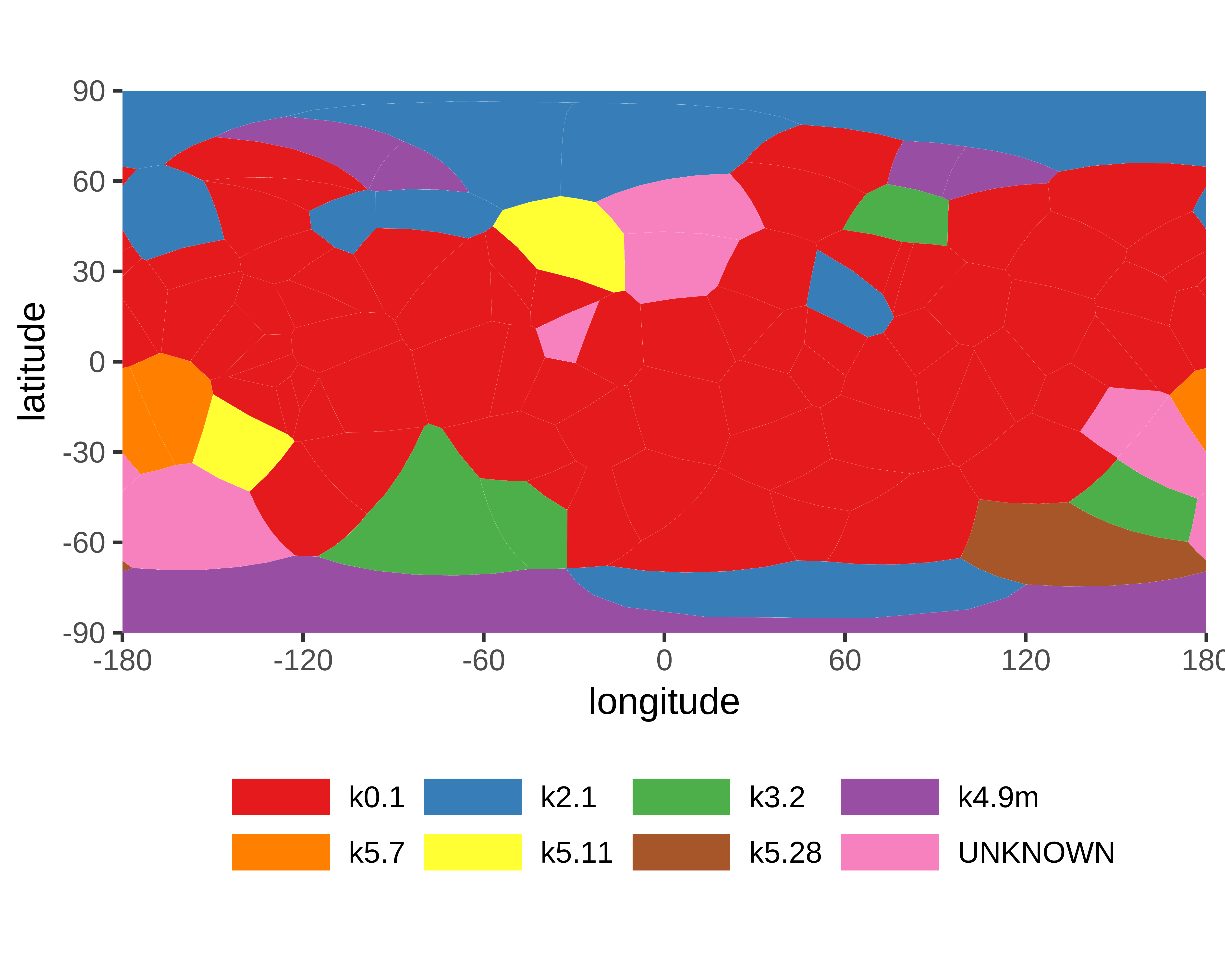
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... His491 <-> Bridging ionZn603 <-> Asp300 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> His346 ... His304 <-> Bridging ionZn603 <-> His491 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> Asp345 ... His304 <-> Bridging ionZn603 <-> His491 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Val549 ... His491 <-> Bridging ionZn603 <-> Asp300 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Cys556 <-> Cys545 ... His491 <-> Bridging ionZn603 <-> Asp300 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Val549 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Cys556 <-> Cys545 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> His346 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn603 <-> His491 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> His346 ... His304 <-> Bridging ionZn603 <-> His491 ... Val549 <-> Bridging ionCa604 <-> Asp553 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Cys556 <-> Cys545 ... His491 <-> Bridging ionZn603 <-> His304 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> Asp345 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn603 <-> His491 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> Asp345 ... His304 <-> Bridging ionZn603 <-> His491 ... Val549 <-> Bridging ionCa604 <-> Asp553 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Cys556 <-> Cys545 ... His491 <-> Bridging ionZn603 <-> His304 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Val549 ... His491 <-> Bridging ionZn603 <-> Asp300 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Val549 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> His346 ... His304 <-> Bridging ionZn603 <-> His491 ... Val549 <-> Bridging ionCa604 <-> Gly551 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> Asp345 ... His304 <-> Bridging ionZn603 <-> His491 ... Val549 <-> Bridging ionCa604 <-> Gly551 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Val549 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Val549 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Val549 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Val549 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Gly551 ... His491 <-> Bridging ionZn603 <-> Asp300 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Gly551 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> His346 ... His304 <-> Bridging ionZn603 <-> His491 ... Gly551 <-> Bridging ionCa604 <-> Asp553 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> Asp345 ... His304 <-> Bridging ionZn603 <-> His491 ... Gly551 <-> Bridging ionCa604 <-> Asp553 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His304 <-> Bridging ionZn603 <-> Asp300 ... Cys126 <-> Cys90 ... Ala31 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... Cys314 <-> Cys231 ... Cys126 <-> Cys90 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Val549 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Cys556 <-> Cys545 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Val549 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Cys556 <-> Cys545 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> His346 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn603 <-> His491 ... Val549 <-> Bridging ionCa604 <-> Asp553 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Cys556 <-> Cys545 ... His491 <-> Bridging ionZn603 <-> His304 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> Asp345 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn603 <-> His491 ... Val549 <-> Bridging ionCa604 <-> Asp553 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Cys556 <-> Cys545 ... His491 <-> Bridging ionZn603 <-> His304 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Val549 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Val549 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> His346 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn603 <-> His491 ... Val549 <-> Bridging ionCa604 <-> Gly551 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> Asp345 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn603 <-> His491 ... Val549 <-> Bridging ionCa604 <-> Gly551 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Val549 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Val549 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Val549 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Val549 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Gly551 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Gly551 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> His346 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn603 <-> His491 ... Gly551 <-> Bridging ionCa604 <-> Asp553 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Ala31 ... Thr89 <-> Bridging ionZn602 <-> Asp345 ... Cys314 <-> Cys231 ... His304 <-> Bridging ionZn603 <-> His491 ... Gly551 <-> Bridging ionCa604 <-> Asp553 ... Chain closureLeu560 <-> Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Asp553 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> Asp300 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Cys231 <-> Cys314 ... His346 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureAla31 <-> Leu560 ... Gly551 <-> Bridging ionCa604 <-> Asp547 ... His491 <-> Bridging ionZn603 <-> His304 ... Cys231 <-> Cys314 ... Asp345 <-> Bridging ionZn602 <-> Thr89 ... Ala31 |
probabilistic |
|

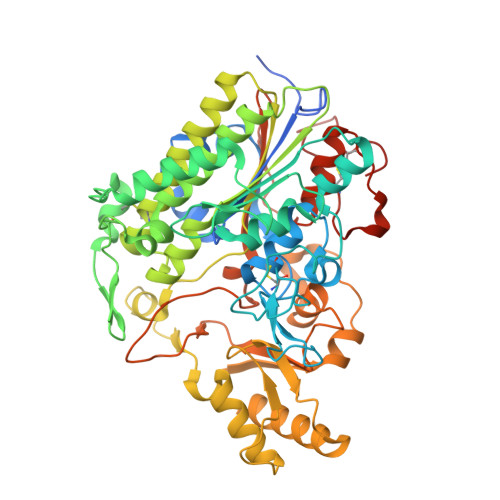
 Genus: 191
Genus: 191