
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
41 | 19-263 | 245 | 1-18, 264-505 | 18 | 242 | knot |
Chain Sequence |
QEVDLTNCDREPIHIPGAIQPHGVLLVLSEPGLVLTHASENAPAVLGNSAEQLLGAPLGHFIEPSVREPLEADLRSARLKQLNPLKVVWRVDGVDRFFDGIAHRHQGRLILELEPSSHREAVPFLSFFHAVRDGLSRLRDARDLQELCEAVVQEVRGLTGFDRAIIYRFDAEWNGSVIAEARDARADPYLGLHFPASDIPRQARELYQLNWLRIIPTIDYQPARVRALPGHGEPLDLSFSVLRSVSPIHLEYLHNMGVQASMSISLMKDGKLWGLISCHQVSGTRYVPYEVRTACEFLGEVMSSLLAAKEGNEDYDQRIRAKSIHAALLERMAREVDFVSGLASQESGLLELVHAHGAAIHFHGRTTVLGQAPSDEALTGLIEWLGSRTGEGVFCTDRLAREYPEAQAFQEVAAGLMAFSMSRGRNNFVLWFRPEAVQTVNWSGNPTKAVEFDQGGPRLHPRKSFELWKETVRGRCLPWKAYEVEAASELRRSIIDVALQRSEEL
|
Knotoid cutoff: 0.5
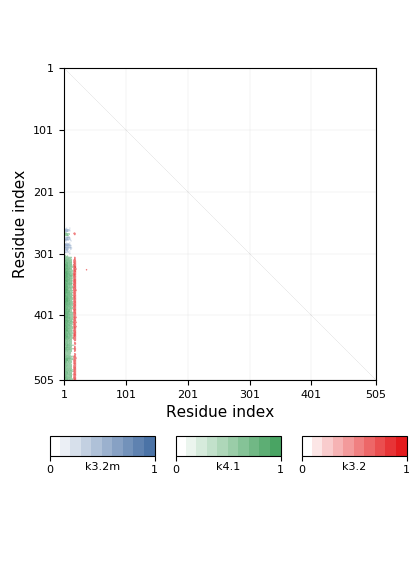
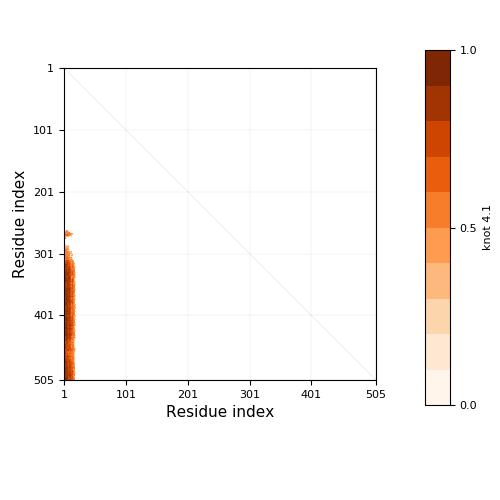
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
4.1 | 17-303 | 287 | 1-16, 304-505 | 16 | 202 | knot | ||||
| view details |
|
3.2m | 17-259 | 243 | 1-16 | 268-505 | 260-267 | 16 | 238 | slipknot | ||
| view details |
|
4.1 | 13-267 | 255 | 1-12 | 274-505 | 268-273 | 12 | 232 | slipknot | ||
| view details |
|
3.2m | 17-268 | 252 | 1-16 | 279-505 | 269-278 | 16 | 227 | slipknot | ||
| view details |
|
3.2m | 15-278 | 264 | 1-14 | 282-505 | 279-281 | 14 | 224 | slipknot | ||
| view details |
|
3.2m | 17-283 | 267 | 1-16 | 304-505 | 284-303 | 16 | 202 | slipknot | ||
| view details |
|
3.2 | 21-300 | 280 | 301-505 | 1-16 | 17-20 | 16 | 204 | slipknot |
| sequence length |
505
|
| structure length |
505
|
| publication title |
Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome
doi rcsb |
| molecule tags |
Signaling protein
|
| molecule keywords |
Photoreceptor-histidine kinase BphP
|
| source organism |
Stigmatella aurantiaca dw4/3-1
|
| total genus |
 Genus: 139 Genus: 139
|
| ec nomenclature | |
| pdb deposition date | 2017-10-14 |
| KnotProt deposition date | 2018-09-25 |
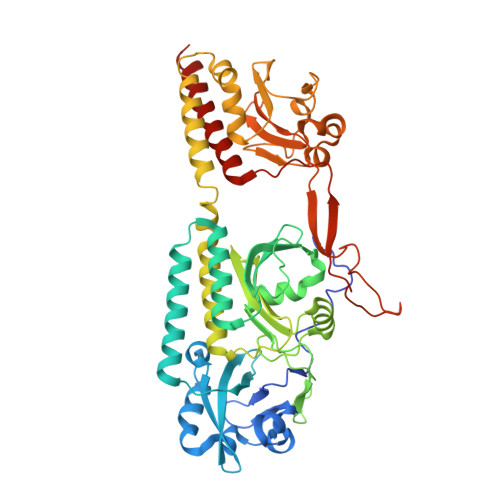
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...