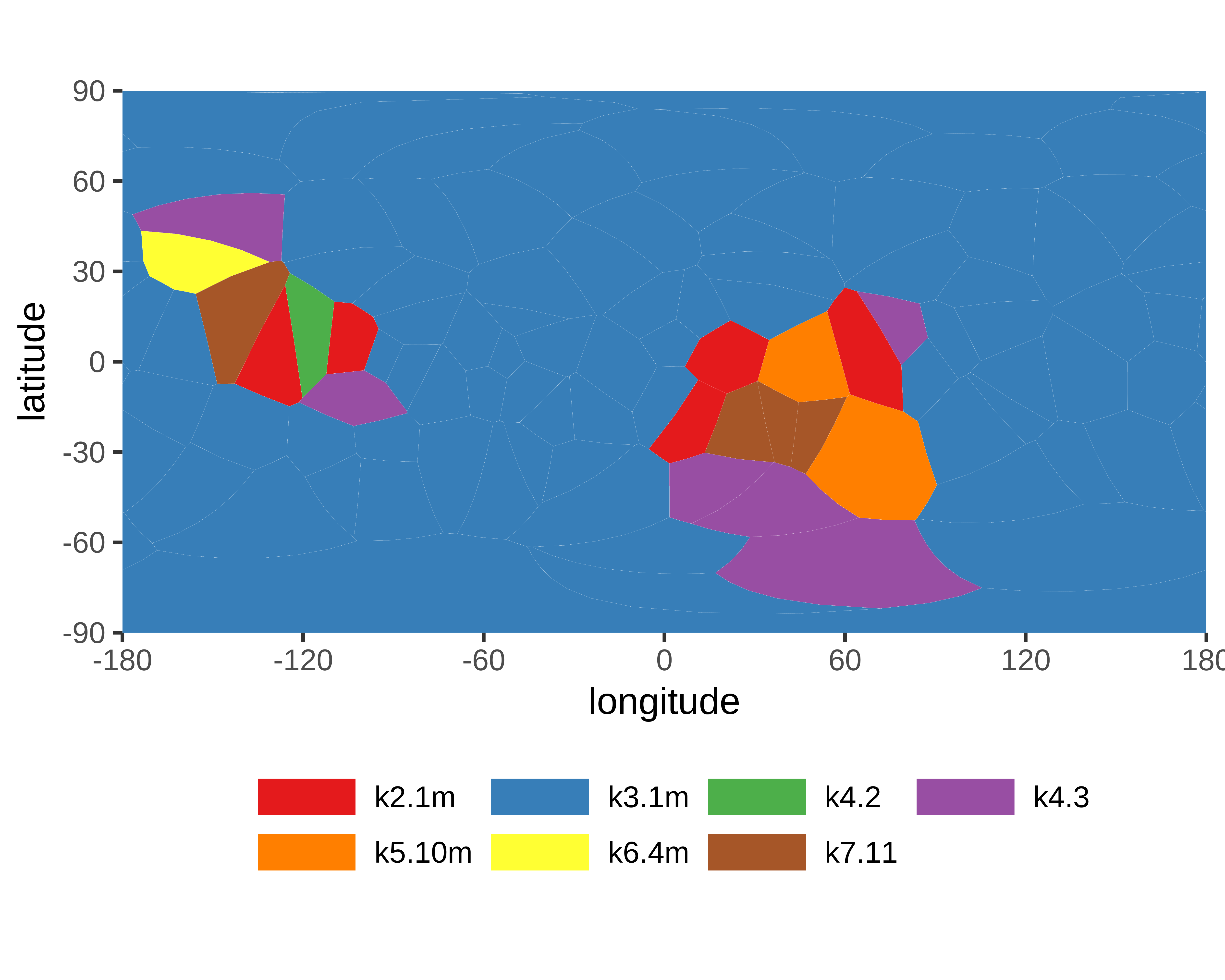
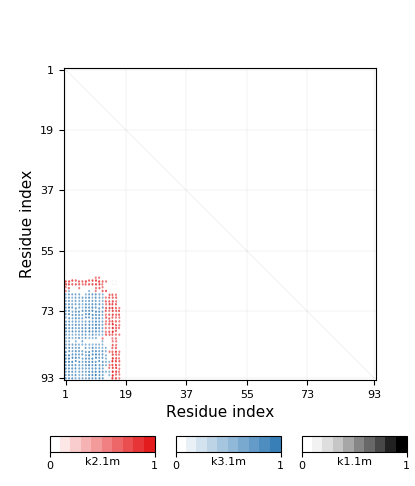
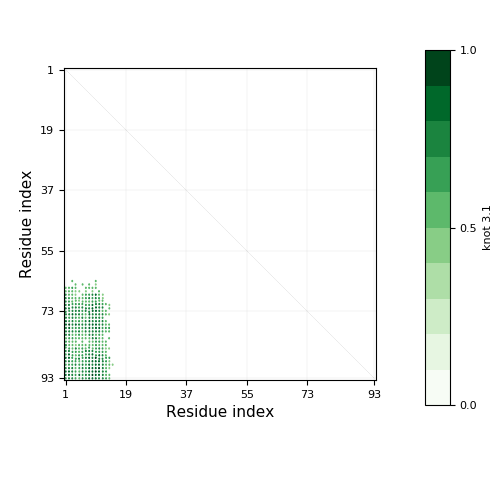
|
|
|
|
K -31
|
Cys11 ... Cys85 <-> Bridging ionZn103 <-> Cys11 |
ion-based |
|
|
|
|
|
K -31
|
Cys11 ... Cys72 <-> Bridging ionZn101 <-> Cys75 ... Cys85 <-> Bridging ionZn103 <-> Cys11 |
ion-based |
|
|
|
|
|
K -31
|
Cys11 ... Cys30 <-> Bridging ionZn101 <-> Cys33 ... Cys85 <-> Bridging ionZn103 <-> Cys11 |
ion-based |
|
|
|
|
|
K -31
|
Cys11 <-> Bridging ionZn103 <-> Cys85 ... Cys61 <-> Bridging ionZn102 <-> Cys58 ... Cys11 |
ion-based |
|
|
|
|
|
K -31
|
Pro6 ... Cys72 <-> Bridging ionZn101 <-> Cys75 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys30 <-> Bridging ionZn101 <-> Cys33 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys58 <-> Bridging ionZn102 <-> Cys61 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys46 <-> Bridging ionZn103 <-> Cys49 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Cys11 <-> Bridging ionZn103 <-> Cys85 ... Cys75 <-> Bridging ionZn101 <-> Cys72 ... Cys61 <-> Bridging ionZn102 <-> Cys58 ... Cys11 |
ion-based |
|
|
|
|
|
K -31
|
Cys11 <-> Bridging ionZn103 <-> Cys85 ... Cys61 <-> Bridging ionZn102 <-> Cys58 ... Cys33 <-> Bridging ionZn101 <-> Cys30 ... Cys11 |
ion-based |
|
|
|
|
|
K -31
|
Pro6 ... Cys58 <-> Bridging ionZn102 <-> Cys61 ... Cys72 <-> Bridging ionZn101 <-> Cys75 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys30 <-> Bridging ionZn101 <-> Cys33 ... Cys58 <-> Bridging ionZn102 <-> Cys61 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys46 <-> Bridging ionZn103 <-> Cys49 ... Cys72 <-> Bridging ionZn101 <-> Cys75 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys30 <-> Bridging ionZn101 <-> Cys33 ... Cys46 <-> Bridging ionZn103 <-> Cys49 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys46 <-> Bridging ionZn103 <-> Cys49 ... Cys58 <-> Bridging ionZn102 <-> Cys61 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys46 <-> Bridging ionZn103 <-> Cys49 ... Cys58 <-> Bridging ionZn102 <-> Cys61 ... Cys72 <-> Bridging ionZn101 <-> Cys75 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|
|
|
|
|
K -31
|
Pro6 ... Cys30 <-> Bridging ionZn101 <-> Cys33 ... Cys46 <-> Bridging ionZn103 <-> Cys49 ... Cys58 <-> Bridging ionZn102 <-> Cys61 ... Chain closureLeu98 <-> Pro6 |
probabilistic |
|

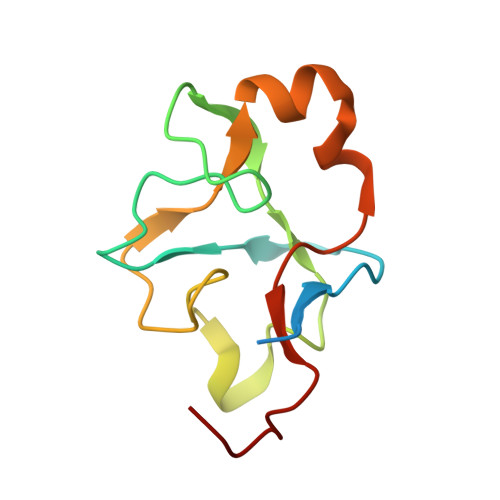
 Genus: 16
Genus: 16