
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 22-257 | 236 | 1-21, 258-260 | 21 | 3 | knot |
Chain Sequence |
SHHWGYGKHNGPEHWHKDFPIAKGERQSPVDIDTHTAKYDPSLKPLSVSYDQATSLRILNNGHAFNVEFDDSQDKAVLKGGPLDGTYRLIQFHFHWGSLDGQGSEHTVDKKKYAAELHLVHWNTKYGDFGKAVQQPDGLAVLGIFLKVGSAKPGLQKVVDVLDSIKTKGKSADFTNFDPRGLLPESLDYWTYPGSLTTPPLLECVTWIVLKEPISVSSEQVLKFRKLNFNGEGEPEELMVDNWRPAQPLKNRQIKASFK
|
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
+ 31 | 26-257 | 232 | 1-25, 258-260 | 25 | 3 | knot |
| Fingerprint | Knot forming loop | Loop type | ||||
|---|---|---|---|---|---|---|
|
|
K +31 | Ser2 <-> Bridging ionCu302 <-> His4 ... Chain closureLys261 <-> Ser2 |
probabilistic | |||
|
|
K +31 | Ser2 ... His94 <-> Bridging ionCu301 <-> His96 ... Chain closureLys261 <-> Ser2 |
probabilistic | |||
|
|
K +31 | Ser2 ... His96 <-> Bridging ionCu301 <-> His119 ... Chain closureLys261 <-> Ser2 |
probabilistic | |||
|
|
K +31 | Ser2 ... His94 <-> Bridging ionCu301 <-> His119 ... Chain closureLys261 <-> Ser2 |
probabilistic | |||
|
|
K +31 | Ser2 <-> Bridging ionCu302 <-> His4 ... His94 <-> Bridging ionCu301 <-> His96 ... Chain closureLys261 <-> Ser2 |
probabilistic | |||
|
|
K +31 | Ser2 <-> Bridging ionCu302 <-> His4 ... His96 <-> Bridging ionCu301 <-> His119 ... Chain closureLys261 <-> Ser2 |
probabilistic | |||
|
|
K +31 | Ser2 <-> Bridging ionCu302 <-> His4 ... His94 <-> Bridging ionCu301 <-> His119 ... Chain closureLys261 <-> Ser2 |
probabilistic |
Chain Sequence |
SHHWGYGKHNGPEHWHKDFPIAKGERQSPVDIDTHTAKYDPSLKPLSVSYDQATSLRILNNGHAFNVEFDDSQDKAVLKGGPLDGTYRLIQFHFHWGSLDGQGSEHTVDKKKYAAELHLVHWNTKYGDFGKAVQQPDGLAVLGIFLKVGSAKPGLQKVVDVLDSIKTKGKSADFTNFDPRGLLPESLDYWTYPGSLTTPPLLECVTWIVLKEPISVSSEQVLKFRKLNFNGEGEPEELMVDNWRPAQPLKNRQIKASFK
|
Knotoid cutoff: 0.5

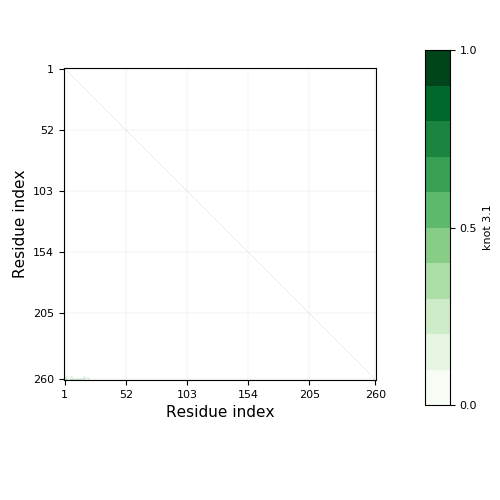
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
3.1 | 28-257 | 230 | 1-27, 258-259 | 27 | 2 | knot | ||||
| view details |
|
2.1 | 26-253 | 228 | 1-25 | 259-259 | 254-258 | 25 | 1 | slipknot |
| sequence length |
259
|
| structure length |
259
|
| publication title |
Structure and mechanism of copper-carbonic anhydrase II: a nitrite reductase.
pubmed doi rcsb |
| molecule tags |
Lyase
|
| molecule keywords |
Carbonic anhydrase 2
|
| source organism |
Homo sapiens
|
| total genus |
 Genus: 75 Genus: 75
|
| ec nomenclature |
ec
4.2.1.1: Alcohol dehydrogenase. |
| pdb deposition date | 2019-06-19 |
| KnotProt deposition date | 2020-03-30 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00194 | Carb_anhydrase | Eukaryotic-type carbonic anhydrase |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...