-
Read more
- Introduction
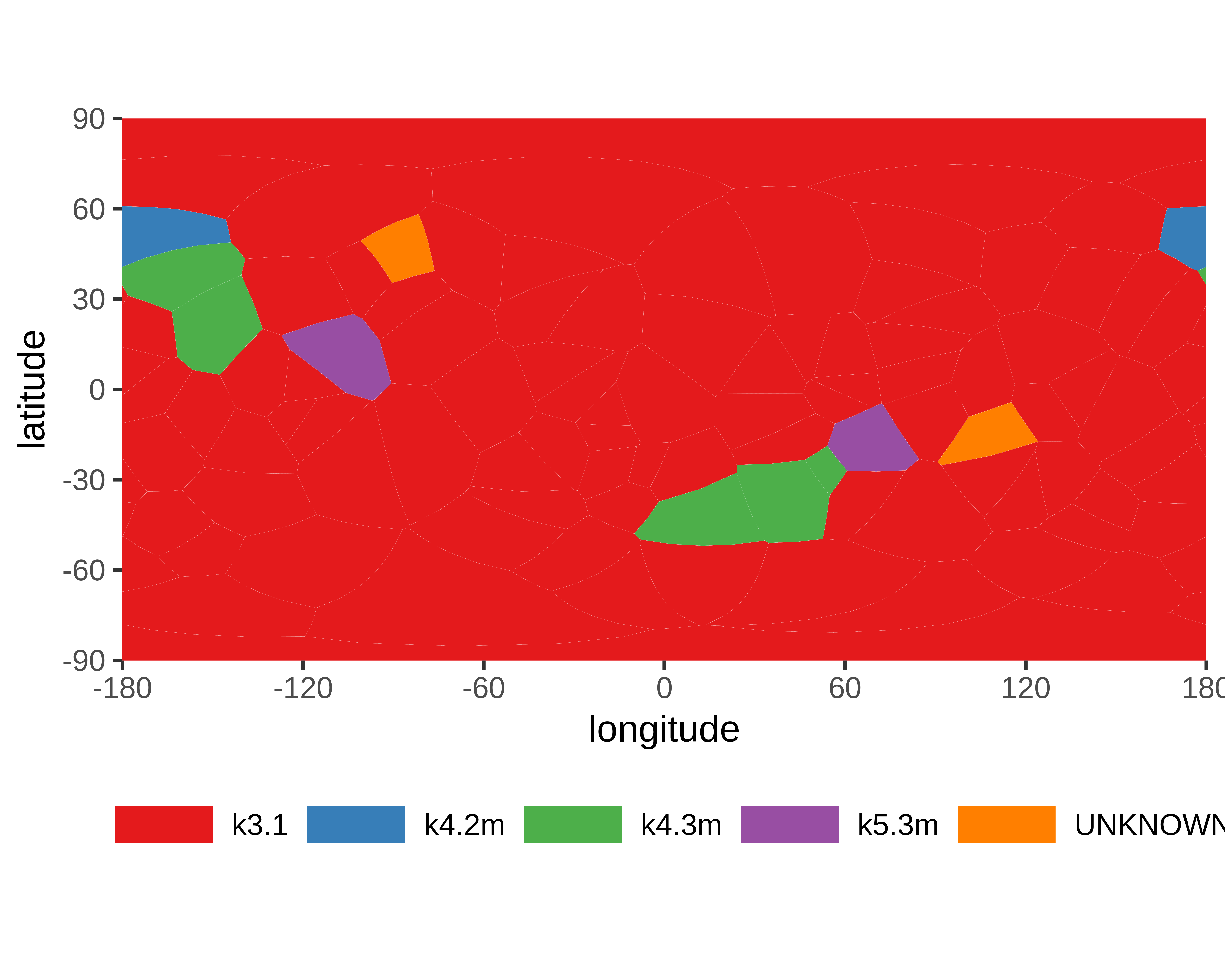
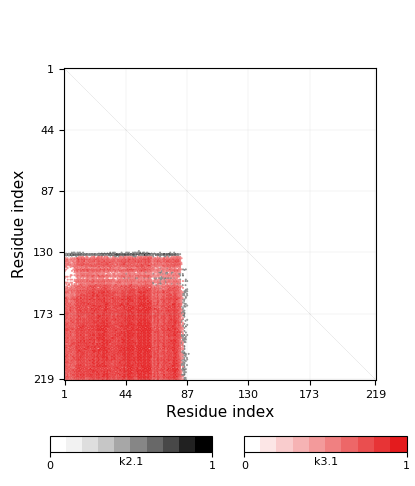
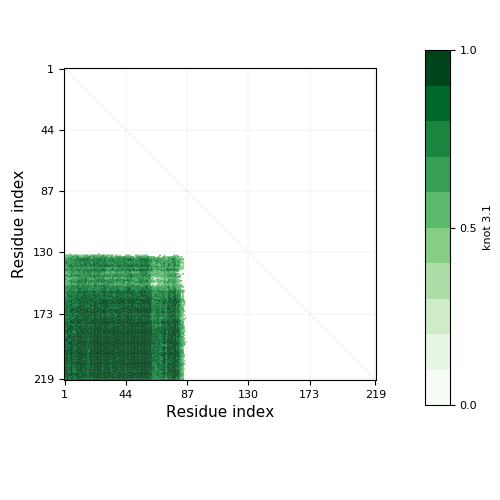
- Knot detection
- Knotoids
- Knots based on dissulfide and ion bonds
- Cysteine knots
- How to
- Search and browse database
- Analyze single structure and trajectory
- Interpret knotting data
- Apply results of fingerprint analysis
- Prepare files to submit data
- About
- List of cysteine knots (non-redundant)
- List of cysteine knots (all)
- Database statistics
- API
- Server status
- KnotProt references
- Contact

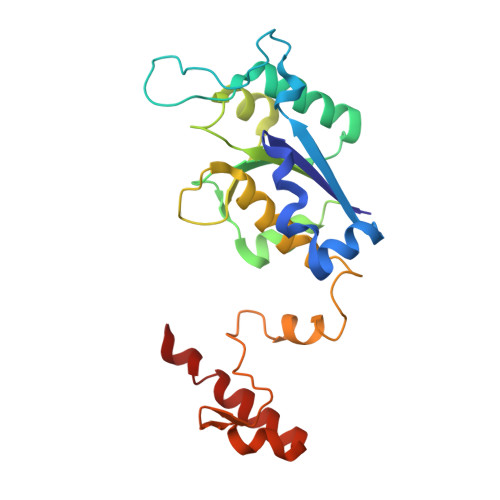
 Genus: 55
Genus: 55