
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
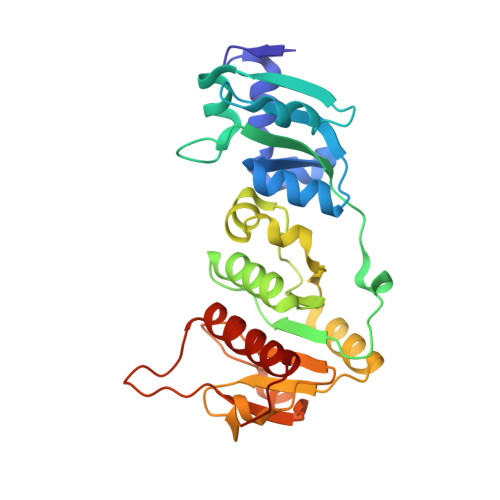
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 185-225 | 41 | 1-184, 226-260 | 184 | 35 | knot |
Chain Sequence |
MRIESPQNPRVKALAALKERKERERTGRFLVEGRREVERALEAGLSLETLLLGPKARPEDRALAGGAEVLELSERALARVSTRENPAQVLGVFRLPRRSLAGVTLGAAPLVLVLLGLEKPGNLGAILRAADGAGADLVLVAEGVDLFSPQVIRNSTGAVFALPVYPVAEGEAARFLEEHNLPLVAATPEGERLYWEGDYRGGVAFLLGAEDKGLPEAWKRRAQVRVRIPMRGRADSLNVAVTAALLLYEALRQRSGGAPL
|
Knotoid cutoff: 0.5

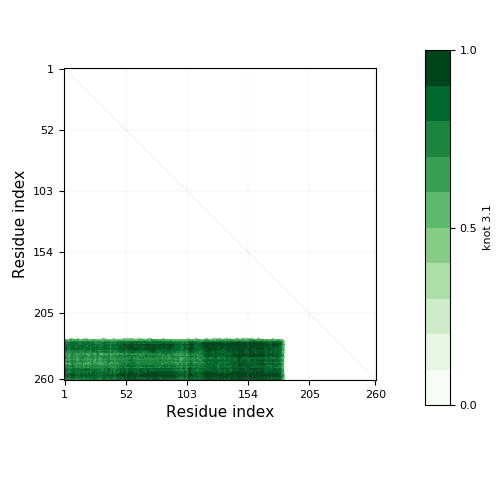
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
3.1 | 186-226 | 41 | 1-185, 227-260 | 185 | 34 | knot |
| sequence length |
260
|
| structure length |
260
|
| publication title |
Crystal Structure of TTHA0275 from Thermus thermophilus (HB8)
rcsb |
| molecule tags |
Transferase
|
| molecule keywords |
RNA 2'-O ribose methyltransferase
|
| source organism |
Thermus thermophilus
|
| total genus |
 Genus: 80 Genus: 80
|
| ec nomenclature | |
| pdb deposition date | 2014-12-01 |
| KnotProt deposition date | 2016-01-22 |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...