Knotting data
The information about topology of a given structure is presented under "Knotting data" and "Detailed topology (knotoids)". The first screen "Knotting data" presents the "knotting fingerprint of a protein" that encodes information about locations of the "knot core", "knot tails", "slipknot loops", and "slipknot tails", whose definitions are given below. The same information is also shown in a graphical (in JSmol) and sequential representation. The topological fingerprint was introduced in [1] and motivated by work [2]; for various applications see also [3]. The second screen "Detailed topology (knotoids)" presents more detailed information, which includes characterization of knotoids (whose type depends on a choice of locations of open ends), whose application in the context of proteins is discussed in [4].
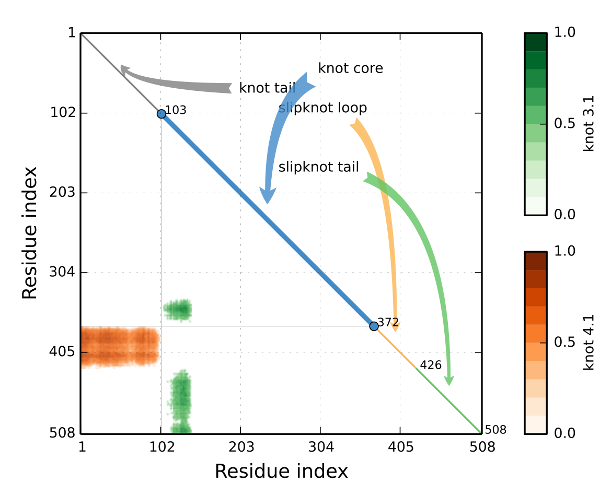
We discuss first the screen "Knotting data", which summarizes information about knots formed by the whole chain and all subchains of a given protein. A knotting fingerprint takes form of a matrix diagram, constructed as follows. For a given chain of length N, all its subchains spanned between amino acids k and l (for 1≤k<l≤N) are analyzed. If a subchain from k to l is knotted (as determined by the analysis described in section Knot detection), then a point with coordinates (k,l) is denoted in relevant color in a plot. For example, point representing knots 31, 41, and 51 are respectively green, red and orange. The intensity of a color represents a probability of detecting a given knot (knots are detected only with some probability, which is a consequence of a choice of its closure to a closed loop, as explained in section Knot detection). The figure below presents an example of a knotting fingerprint, which in this case contains areas representing a knot and two types of slipknots.
In a given knot or slipknot several geometric elements can be distinguished. These elements are denoted in different colors along diagonals of matrix diagrams in database, and also shown schematically in figure below:
- knot core (thick line in figure A below, and in blue in “Knot plot”): the shortest subchain for which a knot is detected (i.e. after cutting an aminoacid from any terminal of such a subchain, just a trivial knot would be detected); note that “knot core” is defined also for a slipknot
- knot tail (thin lines in figure A below, and in grey in “Knot plot”): a segment between one terminal of a knotted chain and its “knot core”
- slipknot tail (thin lines in figures B and C below, and in green in “Knot plot”): in a structure with a slipknot (i.e. a configuration in which the whole chain is unknotted, but it has a knotted subchain), the longest segment starting at one terminal, for which no change in topology is detected
- slipknot loop (dashed lines in figures B and C below, and in yellow in “Knot plot”): in a structure with a slipknot, a segment between a “slipknot tail” and a “knot core”
Frequently knotting fingerprints (matrices encoding the knotting types of all subchains) show the presence of more than one knot type or the same knot type appears in disjoint territories of the fingerprint. This feature was used to define a topological notation for knotted proteins. In this notation we list distinct knotted territories, ordering them according to the size of the subchain forming a given knot, starting with the largest one. If the entire protein is knotted its topological notation starts with K; if it is unknotted but contains a slipknot its topological notation starts with S. Among the complete protein structures deposited in PDB we found proteins with the following topological notations:
- knots: K31, K4131, K4141, K523131, K61614131
- slipknots: S31, S3131, S313131, S31313131, S314131, S3141313131
Interestingly, the topological notation is strongly conserved among ortologous proteins even if their structure highly diverged during hundreds of million of years from their evolutionarily separation. Therefore topological notation can be used to identify proteins with the same or similar function, as a template to model new proteins (e.g. to impose topological constraints on threading), or to identify new members of a given family, etc.
In the KnotProt we also list (as “putative notations”) a few cases where the notation is associated to protein chains with undetermined fragments and where the “missing” portion was replaced by a line segment. In such cases the line segment can pierce through the existing portion of the chain and introduce spatial structure that is an artifact. In particular, we found cases with topological notations K5131, S715131. At present we do not think that these notations reflect the notation of the entire protein structure of the respective proteins. Once complete structural information about these proteins is provided (e.g. by new crystallization results, or proper reconstruction), the KnotProt analysis will be repeated and the results will be updated.
The screen "Detailed information (knotoids)" presents refined information, which takes into account location of protein’s ends, and identifies a given protein structure as a knotoid. In this screen two graphs are presented. The first one, called “Whole chain analysis”, presents projection globes/maps. The second one presents a generalized fingerprint matrix, which determines the most likely knotoid type for each subchain of the whole chain. Examples of these graphs are shown below in Figures 2 and 3. More details about these two representations are explained at http://knotprot.cent.uw.edu.pl/help_knotoids
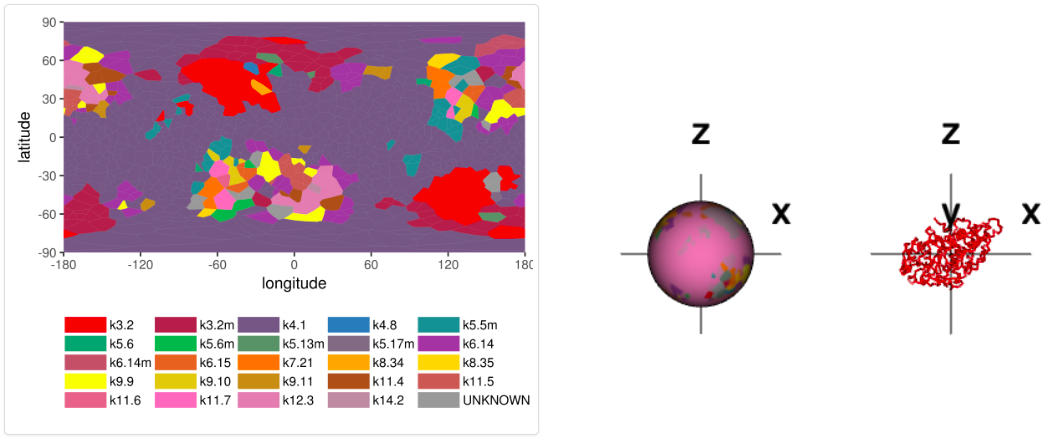
Figure 2. Projection globe/map representing various knotoids obtained from various projections for 1yrl protein.

Figure 3. Refined fingerprint with various knotoids detected for 1yrl protein.
[1] Sulkowska JI, Rawdon EJ, Millett KC, Onuchic JN and Stasiak A (2012)
Conservation of complex knotting and slipknotting patterns in proteins, PNAS 109, E1715–E1723
[2] King NP, Yeates EO, Yeates TO (2007).
Identification of rare slipknots in proteins and their implications for stability and folding, J. Mol. Biol. 373, 153-66.
[3] Millett KC, Rawdon EJ, Stasiak A, and Sułkowska JI (2012)
Identifying knots in proteins, Biochem. Soc. Trans. 41, 533–537.
[4] Goundaroulis D, Gügümcü N, Lambropoulou S, Dorier J, Stasiak A, Kauffman LH (2017)
Topological Models for Open-Knotted Protein Chains Using the Concepts of Knotoids and Bonded Knotoids, Polymers, 9, 444.