
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]

| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 168-213 | 46 | 1-167, 214-240 | 167 | 27 | knot |
Chain Sequence |
PPPALPQLVAEQHVPIPPNDKDTKRLIVVLSNASLETYK--------------YTLLNSDEHIGIMRKMNRDISDARPDITHQCLLTLLDSPINKAGKLQIYIQTAKGVLIEVSPTVRIPRTFKRFAGLMVQLLHRLSIKGTNTNEKLLKVIQNPITDHLPPNCRKVTLSFDAPLVRVRDYVDTLGPNESICVFVGAMAKGPDNFADAYVDEKISISNYSLSASVACSKFCHACEDAWDI
|
Knotoid cutoff: 0.5
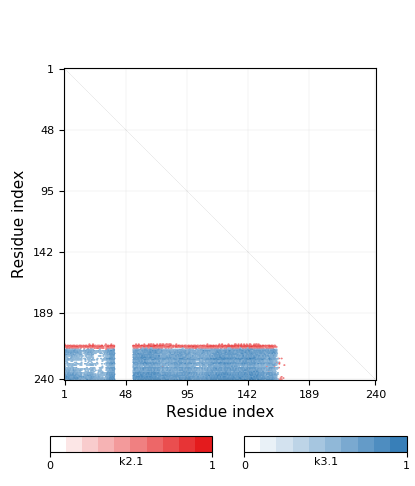
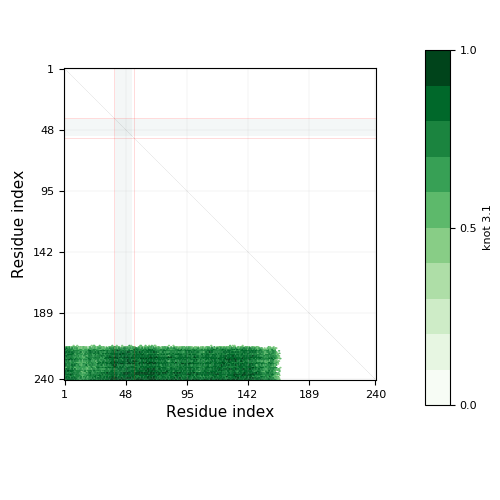
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
3.1 | 168-215 | 48 | 1-167, 216-240 | 167 | 25 | knot | ||||
| view details |
|
2.1 | 165-212 | 48 | 1-164 | 218-240 | 213-217 | 164 | 23 | slipknot |
| sequence length |
240
|
| structure length |
226
|
| publication title |
Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
pubmed doi rcsb |
| molecule tags |
Ribosome
|
| molecule keywords |
Periodic tryptophan protein 2-like protein
|
| missing residues |
40-53
|
| total genus |
 Genus: 14 Genus: 14
|
| ec nomenclature | |
| pdb deposition date | 2019-06-10 |
| KnotProt deposition date | 2019-09-07 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| CN | PF03587 | EMG1 | EMG1/NEP1 methyltransferase |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...