|
|
|
|
K +31
|
Asp395 ... Cys481 <-> Cys675 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 |
ion-based |
|
|
|
|
|
K +31
|
Asp395 ... Cys481 <-> Cys675 ... Cys587 <-> Cys573 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 |
ion-based |
|
|
|
|
|
K +31
|
Asp395 ... Cys481 <-> Cys675 ... Cys630 <-> Cys625 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 |
ion-based |
|
|
|
|
|
K +31
|
Asp395 ... Cys481 <-> Cys675 ... Cys630 <-> Cys625 ... Cys587 <-> Cys573 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 |
ion-based |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys587 <-> Cys573 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys630 <-> Cys625 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Cys371 <-> Cys358 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Cys380 <-> Cys348 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys630 <-> Cys625 ... Cys587 <-> Cys573 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys587 <-> Cys573 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Cys371 <-> Cys358 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys630 <-> Cys625 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Cys371 <-> Cys358 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys587 <-> Cys573 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Cys380 <-> Cys348 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys630 <-> Cys625 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Cys380 <-> Cys348 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys630 <-> Cys625 ... Cys587 <-> Cys573 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Cys371 <-> Cys358 ... Tyr342 |
probabilistic |
|
|
|
|
|
K +31
|
Chain closureTyr342 <-> Phe686 ... Cys684 <-> Cys405 ... Cys481 <-> Cys675 ... Cys630 <-> Cys625 ... Cys587 <-> Cys573 ... Tyr526 <-> Bridging ionFe690 <-> Asp395 ... Cys380 <-> Cys348 ... Tyr342 |
probabilistic |
|

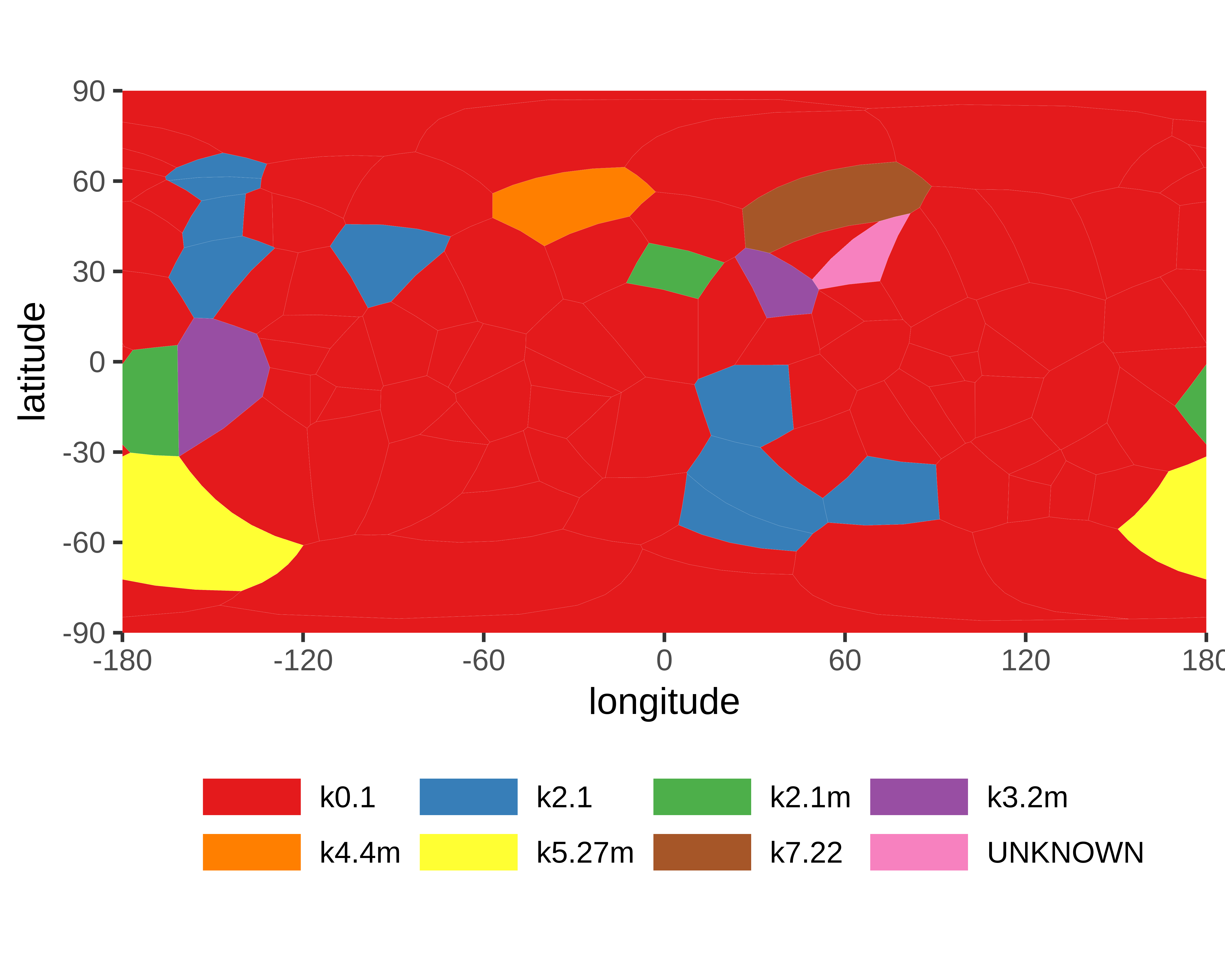

 Genus: 108
Genus: 108