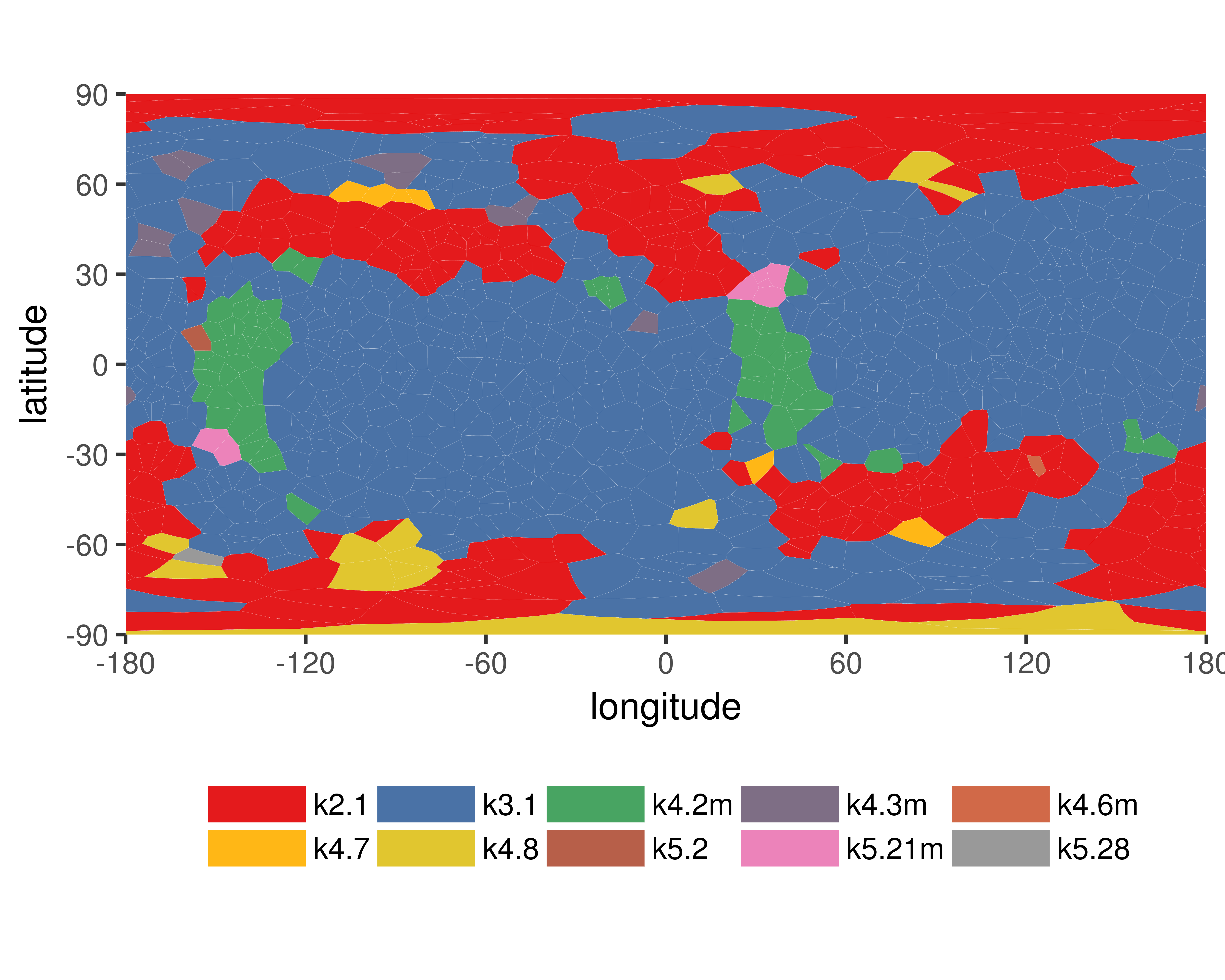
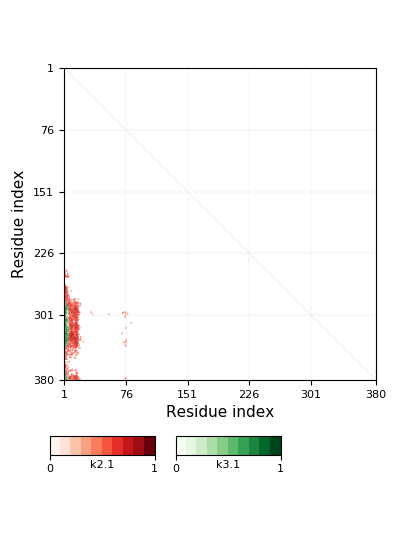
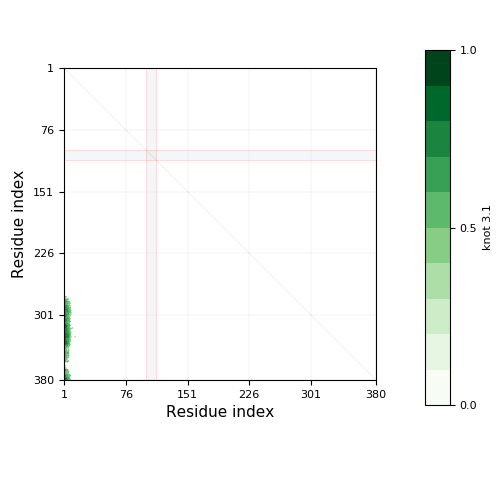
Warning
- Chain breaks within knot 31 (displayed as a gray area on the plot and as '-' on the sequence). The broken part of the chain has been replaced by a straight segment, which may affect what knot types are detected - be careful with interpreting results.

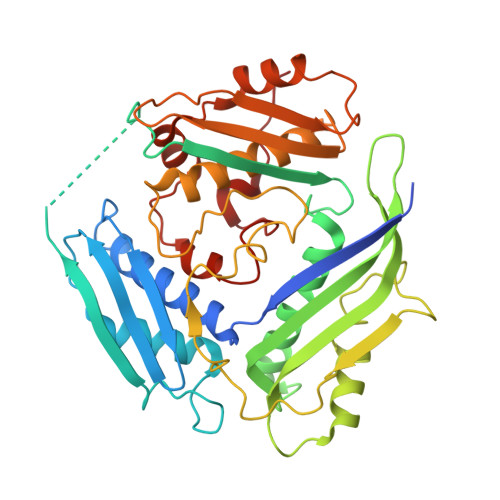
 Genus: 119
Genus: 119