
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 15-116 | 102 | 1-12, 298-303 | 13-14, 117-297 | 12 | 6 | slipknot |
Chain Sequence |
GPAHLPYGGIPTFARAPLVQPDGDWQADVAALGVPFDIALGFRPGARFAPRALREASLRSVPPFTGLDGKTRLQGVTFADAGDVILPSLEPQLAHDRITEAARQVRGRCRVPVFLGGDHSVSYPLLRAFADVPDLHVVQLDAHLDFTDTRNDTKWSNSSPFRRACEALPNLVHITTVGLRGLRFDPEAVAAARARGHTIIPMDDVTADLAGVLAQLPRGQNVYFSVDVDGFDPAVIPGTSSPEPDGLTYAQGMKILAAAAANNTVVGLDLVELAPNLDPTGRSELLMARLVMETLCEVFDHVL
|
Knotoid cutoff: 0.5
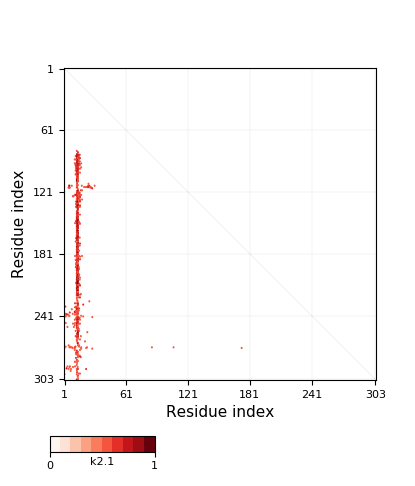
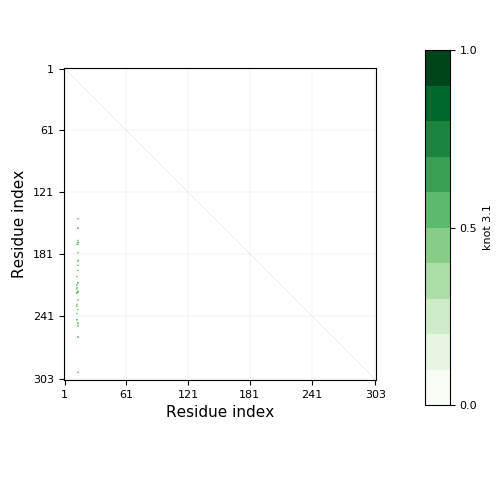
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
2.1 | 16-231 | 216 | 1-10, 241-303 | 11-15, 232-240 | 10 | 63 | slipknot | |||
| view details |
|
2.1 | 17-116 | 100 | 1-11, 137-303 | 12-16, 117-136 | 11 | 167 | slipknot |
| sequence length |
303
|
| structure length |
303
|
| publication title |
Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
pubmed doi rcsb |
| molecule tags |
Hydrolase
|
| molecule keywords |
agmatinase
|
| source organism |
Deinococcus radiodurans
|
| total genus |
 Genus: 114 Genus: 114
|
| ec nomenclature |
ec
3.5.3.11: Agmatinase. |
| pdb deposition date | 2004-08-18 |
| KnotProt deposition date | 2015-02-05 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00491 | Arginase | Arginase family |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Alpha Beta | 3-Layer(aba) Sandwich | Arginase; Chain A | Arginase; Chain A |
#chains in the KnotProt database with same CATH superfamily 4DZ4 A; 1WOH A; 1GQ6 A; 1GQ7 A; 1WOG A; 1WOI A; #chains in the KnotProt database with same CATH topology 4DZ4 A; 1WOH A; 1GQ6 A; 1GQ7 A; 1WOG A; 1WOI A; #chains in the KnotProt database with same CATH homology 4DZ4 A; 1WOH A; 1GQ6 A; 1GQ7 A; 1WOG A; 1WOI A;
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...