|
|
|
|
K -31
|
Asp5 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Asp5 |
ion-based |
|
|
|
|
|
K -31
|
Asp5 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Asp5 |
ion-based |
|
|
|
|
|
K -31
|
Asp5 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Asp5 |
ion-based |
|
|
|
|
|
K -31
|
Asp5 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asp5 |
ion-based |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Val179 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Tyr176 <-> Bridging ionNa702 <-> Val179 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp65 <-> Bridging ionCa703 <-> Asp63 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Asn86 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Ile90 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Trp1 ... Pro47 <-> Bridging ionCa704 <-> Glu83 ... Asp63 <-> Bridging ionCa703 <-> Asp58 ... Asp48 <-> Bridging ionCa701 <-> Thr88 ... Tyr176 <-> Bridging ionNa702 <-> Asp202 ... Chain closureTyr280 <-> Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Val179 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Val179 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp63 <-> Bridging ionCa703 <-> Asp65 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Asn86 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Ile90 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|
|
|
|
|
K -31
|
Chain closureTrp1 <-> Tyr280 ... Asp202 <-> Bridging ionNa702 <-> Tyr176 ... Cys139 <-> Cys137 ... Thr88 <-> Bridging ionCa701 <-> Asp48 ... Asp58 <-> Bridging ionCa703 <-> Asp63 ... Glu83 <-> Bridging ionCa704 <-> Pro47 ... Trp1 |
probabilistic |
|

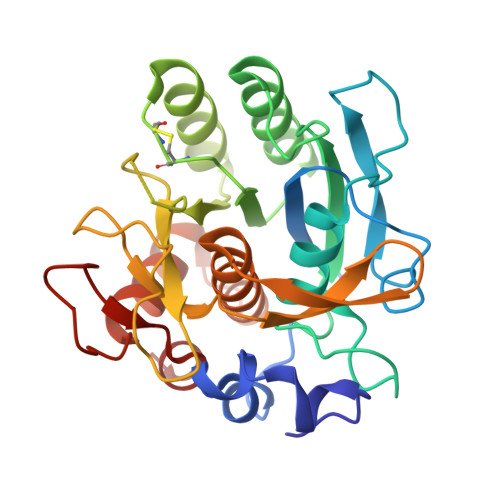
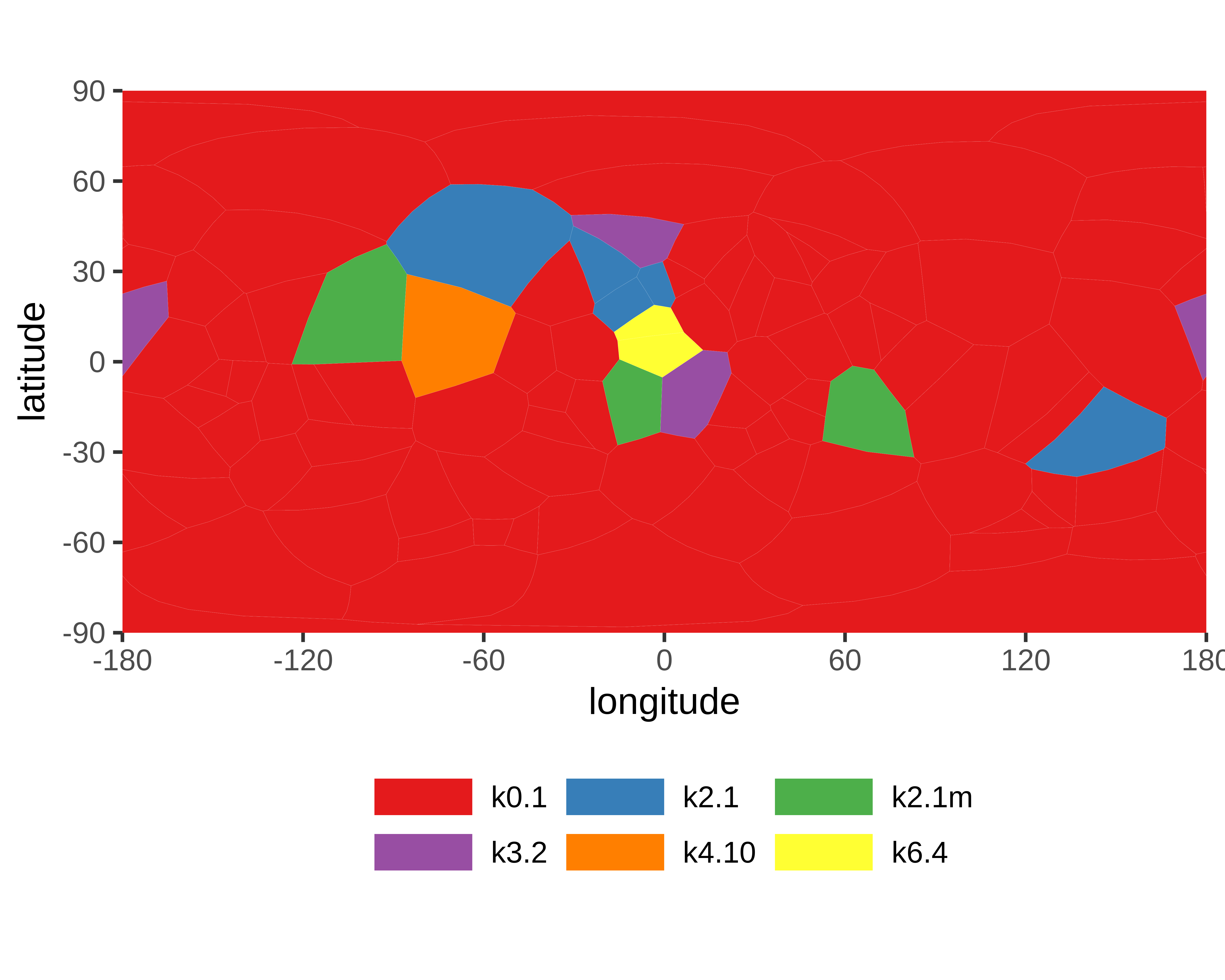

 Genus: 106
Genus: 106