
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 174-251 | 78 | 1-173, 252-332 | 173 | 81 | knot |
Chain Sequence |
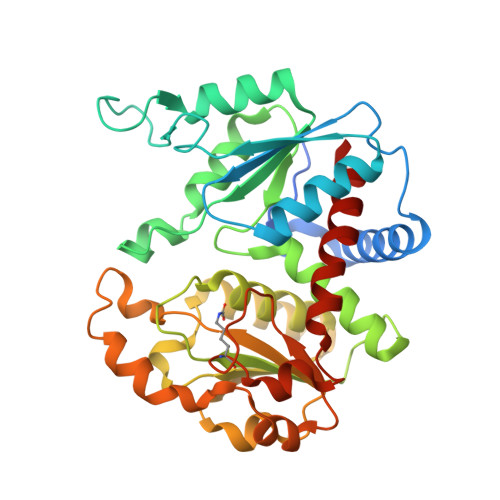
LKHFLNTQDWSRAELDALLTQAALFKRNKLGSELKGKSIALVFFNPSMRTRTSFELGAFQLGGHAVVLQPGKDAWPIEFNLGTVMDGDTEEHIAEVARVLGRYVDLIGVRAFPKFVDWSKDREDQVLKSFAKYSPVPVINMETITHPCQELAHALALQEHFGTPDLRGKKYVLTWTYHPKPLNTAVANSALTIATRMGMDVTLLCPTPDYILDERYMDWAAQNVAESGGSLQVSHDIDSAYAGADVVYAKSWGALPFFGNWEPEKPIRDQYQHFIVDERKMALTNNGVFSHCLPLRRNVKATDAVMDSPNCIAIDEAENRLHVQKAIMAALV
|
Knotoid cutoff: 0.5
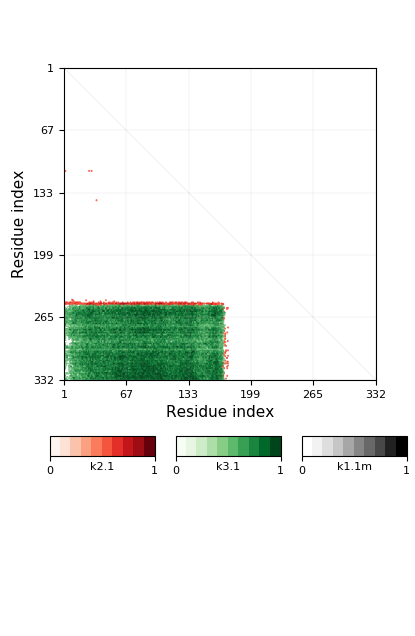
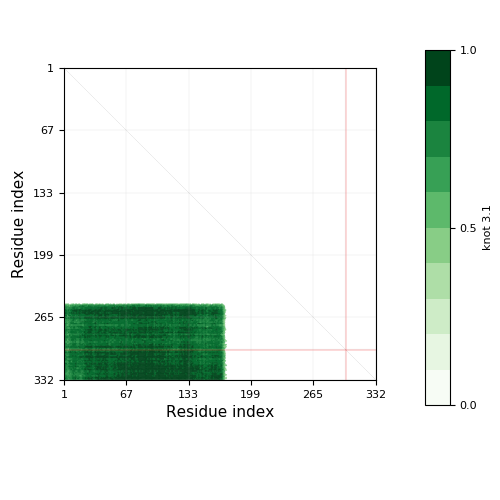
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
3.1 | 174-251 | 78 | 1-173, 252-332 | 173 | 81 | knot | ||||
| view details |
|
2.1 | 131-248 | 118 | 1-3, 252-332 | 4-130, 249-251 | 3 | 81 | slipknot | |||
| view details |
|
2.1 | 140-249 | 110 | 1-130, 252-332 | 131-139, 250-251 | 130 | 81 | slipknot | |||
| view details |
|
2.1 | 171-249 | 79 | 1-139, 252-332 | 140-170, 250-251 | 139 | 81 | slipknot | |||
| view details |
|
3.1 | 175-279 | 105 | 1-173, 285-332 | 174-174, 280-284 | 173 | 48 | slipknot |
| sequence length |
332
|
| structure length |
332
|
| publication title |
Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
pubmed doi rcsb |
| molecule tags |
Transferase
|
| molecule keywords |
N-acetylornithine carbamoyltransferase
|
| source organism |
Xanthomonas campestris pv. campestris
|
| total genus |
 Genus: 125 Genus: 125
|
| ec nomenclature |
ec
2.1.3.9: N-acetylornithine carbamoyltransferase. |
| pdb deposition date | 2010-03-11 |
| KnotProt deposition date | 2014-07-31 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00185 | OTCace | Aspartate/ornithine carbamoyltransferase, Asp/Orn binding domain |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Alpha Beta | 3-Layer(aba) Sandwich | Rossmann fold | Rossmann fold | ||
| Alpha Beta | 3-Layer(aba) Sandwich | Rossmann fold | Rossmann fold |
#chains in the KnotProt database with same CATH superfamily 3M5C A; 3KZK A; 3L04 A; 3KZN A; 3KZC A; 1JS1 X; 3L06 A; 3M5D A; 3KZM A; 3L05 A; 3L02 A; 2FG6 C; 3KZO A; 1YH1 A; 3M4J A; 2G7M C; 3M4N A; #chains in the KnotProt database with same CATH topology 1DBI A; 1P7C A; 1KI2 A; 1P6X A; 3KZK A; 1YRL A; 1E2P A; 1VTK A; 3KZC A; 1JS1 X; 3M5D A; 4IVQ A; 1P75 A; 3KZO A; 4JBX A; 3F0T A; 4IVP A; 3RDP A; 1E2N A; 1E2K A; 1E2I A; 1KI4 A; 1KI7 A; 1F48 A; 3L05 A; 1KI6 A; 3M4J A; 4JBY A; 1OF1 A; 2XTJ A; 3M5C A; 4IVR A; 1LNS A; 1E2J A; 1E2L A; 3L04 A; 1P72 A; 3KZM A; 1II9 A; 3L06 A; 3FR7 A; 1KIM A; 1YVE I; 1QMG A; 1E2H A; 1OSN A; 2G7M C; 3FSG A; 1KI8 A; 3M4N A; 1R4N B; 3KZN A; 1GKU B; 1E2M A; 1P73 A; 3FR8 A; 2KI5 A; 1KI6 B; 1QHI A; 2FG6 C; 3L02 A; 1YH1 A; 1KI3 A; 1R4M B; 2VTK A; 3VTK A; #chains in the KnotProt database with same CATH homology 1DBI A; 3KZK A; 3KZC A; 1JS1 X; 3M5D A; 3KZO A; 3L05 A; 3M4J A; 2XTJ A; 3M5C A; 1LNS A; 3L04 A; 3KZM A; 3L06 A; 2G7M C; 3FSG A; 3M4N A; 3KZN A; 1GKU B; 3L02 A; 2FG6 C; 1YH1 A;
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...