
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 82-216 | 135 | 1-1, 245-267 | 2-81, 217-244 | 1 | 23 | slipknot |
Chain Sequence |
MKEYLTRSNISYFSIGSGTPIIFLHGLSLDKQSTCLFFEPLSNVGQYQRIYLDLPGMGNSDPISPSTSDNVLETLIEAIEEIIGARRFILYGHSYGGYLAQAIAFHLKDQTLGVFLTCPVITADHSKRLTGKHINILEEDINPVENKEYFADFLSMNVIINNQAWHDYQNLIIPGLQKEDKTFIDQLQNNYSFTFEEKLKNINYQFPFKIMVGRNDQVVGYQEQLKLINHNENGEIVLLNRTGHNLMIDQREAVGFHFDLFLDELNS
|
Knotoid cutoff: 0.5
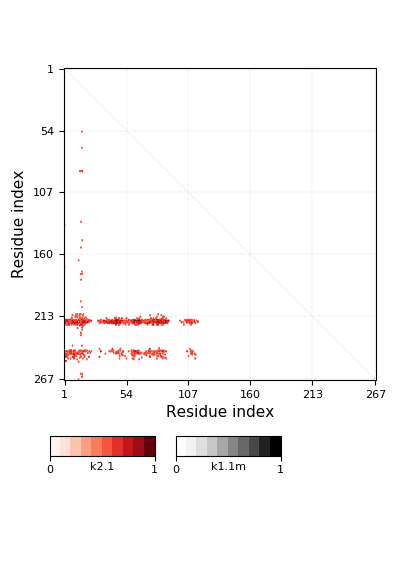

Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
|
2.1 | 24-213 | 190 | 1-23 | 222-267 | 214-221 | 23 | 46 | slipknot | |
| view details |
|
|
2.1 | 24-241 | 218 | 1-23 | 250-267 | 242-249 | 23 | 18 | slipknot | |
| view details |
|
2.1 | 91-213 | 123 | 1-29, 221-267 | 30-90, 214-220 | 29 | 47 | slipknot | |||
| view details |
|
2.1 | 88-241 | 154 | 1-61, 249-267 | 62-87, 242-248 | 61 | 19 | slipknot |
| sequence length |
267
|
| structure length |
267
|
| publication title |
Crystal structure of alpha/beta superfamily hydrolase from Oenococcus oeni PSU-1
rcsb |
| molecule tags |
Hydrolase
|
| molecule keywords |
Alpha/beta superfamily hydrolase
|
| source organism |
Oenococcus oeni
|
| total genus |
 Genus: 83 Genus: 83
|
| ec nomenclature | |
| pdb deposition date | 2009-01-09 |
| KnotProt deposition date | 2014-07-31 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF12697 | Abhydrolase_6 | Alpha/beta hydrolase family |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Alpha Beta | 3-Layer(aba) Sandwich | Rossmann fold | Rossmann fold |
#chains in the KnotProt database with same CATH superfamily 1LNS A; 3FSG A; #chains in the KnotProt database with same CATH topology 3RDP A; 3L04 A; 1E2H A; 1VTK A; 1YRL A; 3L02 A; 3M5C A; 1KI8 A; 2XTJ A; 1E2J A; 2G7M C; 1P7C A; 1KIM A; 1YVE I; 3F0T A; 1E2P A; 1KI6 A; 4IVR A; 1KI3 A; 1P72 A; 3KZN A; 3M5D A; 1E2N A; 1P73 A; 1R4M B; 1JS1 X; 3FR7 A; 3KZM A; 1E2I A; 1GKU B; 3KZO A; 4JBX A; 1KI6 B; 1E2L A; 1E2M A; 1KI7 A; 2VTK A; 1P75 A; 3L06 A; 2KI5 A; 1YH1 A; 3FR8 A; 3M4N A; 4JBY A; 4IVQ A; 1E2K A; 1OF1 A; 3M4J A; 1II9 A; 1KI4 A; 1KI2 A; 1P6X A; 1R4N B; 1QMG A; 4IVP A; 3L05 A; 1QHI A; 2FG6 C; 3KZC A; 3KZK A; 1OSN A; 1DBI A; 1LNS A; 3FSG A; 3VTK A; 1F48 A; #chains in the KnotProt database with same CATH homology 3L04 A; 3M5C A; 3L02 A; 2XTJ A; 2G7M C; 3FSG A; 3KZN A; 3M5D A; 1JS1 X; 3KZM A; 1GKU B; 3KZO A; 3L06 A; 1YH1 A; 3M4N A; 3M4J A; 3L05 A; 2FG6 C; 3KZK A; 1DBI A; 1LNS A; 3KZC A;
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...