
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 77-119 | 43 | 1-76, 120-156 | 76 | 37 | knot |
Chain Sequence |
MLDIVLYEPEIPQNTGNIIRLCANTGFRLHLIEPLGFTWDDKRLRRSGLDYHEFAEIKRHKTFEAFLESEKPKRLFALTTKGCPAHSQVKFKLGDYLMFGPETRGIPMSILNEMPMEQKIRIPMTANSRSMNLSNSVAVTVYEAWRQLGYKGAVNL
|
Knotoid cutoff: 0.5
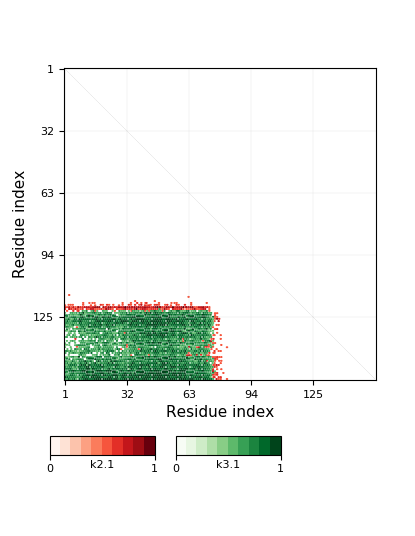
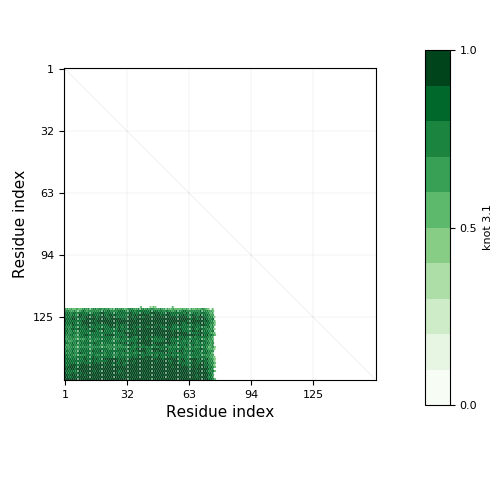
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
3.1 | 77-121 | 45 | 1-76, 122-156 | 76 | 35 | knot | ||||
| view details |
|
2.1 | 76-117 | 42 | 1-75 | 123-156 | 118-122 | 75 | 34 | slipknot | ||
| view details |
|
3.1 | 79-138 | 60 | 139-156 | 1-76 | 77-78 | 76 | 17 | slipknot | ||
| view details |
|
3.1 | 79-129 | 51 | 1-76, 137-156 | 77-78, 130-136 | 76 | 20 | slipknot | |||
| view details |
|
2.1 | 79-136 | 58 | 1-76, 138-156 | 77-78, 137-137 | 76 | 19 | slipknot | |||
| view details |
|
2.1 | 80-138 | 59 | 139-156 | 1-78 | 79-79 | 78 | 17 | slipknot |
| sequence length |
156
|
| structure length |
156
|
| publication title |
Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a Cofactor Bound at a Site Formed by a Knot
pubmed doi rcsb |
| molecule tags |
Transferase
|
| molecule keywords |
Hypothetical tRNA/rRNA methyltransferase HI0766
|
| source organism |
Haemophilus influenzae
|
| total genus |
 Genus: 45 Genus: 45
|
| ec nomenclature |
ec
2.1.1.207: tRNA (cytidine(34)-2'-O)-methyltransferase. |
| pdb deposition date | 2002-10-02 |
| KnotProt deposition date | 2014-07-31 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00588 | SpoU_methylase | SpoU rRNA Methylase family |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Alpha Beta | 3-Layer(aba) Sandwich | Alpha/beta knot | Alpha/beta knot |
#chains in the KnotProt database with same CATH superfamily 3DCM X; 4CNG B; 1X7P A; 1VHK A; 1MXI A; 2O3A A; 2YY8 A; 4E8B A; 2CX8 A; 1NS5 A; 4H3Y A; 4IG6 A; 1X7O A; 3IEF A; 3ILK A; 1UAK A; 3N4K A; 1K3R A; 4FAK A; 1O6D A; 4CNF B; 2HA8 A; 1NXZ A; 1VHY A; 3AXZ A; 1ZJR A; 4CNE B; 3IC6 A; 3NK6 A; 3L8U A; 3ONP A; 3NK7 A; 3AI9 X; 3KY7 A; 1UAL A; 1UAM A; 3AIA A; 1GZ0 A; 3N4J A; 4H3Z A; 1J85 A; 3GYQ A; 1V6Z A; 1P9P A; 1V2X A; 2Z0Y A; 2I6D A; 2QWV A; 3KTY A; 3KNU A; 3QUV A; 3E5Y A; 1VH0 A; 1TO0 A; 2QMM A; 1UAJ A; #chains in the KnotProt database with same CATH topology 3DCM X; 4CNG B; 1X7P A; 1VHK A; 1MXI A; 2O3A A; 2YY8 A; 4E8B A; 2CX8 A; 1NS5 A; 4H3Y A; 4IG6 A; 1X7O A; 3IEF A; 3ILK A; 1UAK A; 3N4K A; 1K3R A; 4FAK A; 1O6D A; 4CNF B; 2HA8 A; 1NXZ A; 1VHY A; 3AXZ A; 1ZJR A; 4CNE B; 3IC6 A; 3NK6 A; 3L8U A; 3ONP A; 3NK7 A; 3AI9 X; 3KY7 A; 1UAL A; 1UAM A; 3AIA A; 1GZ0 A; 3N4J A; 4H3Z A; 1J85 A; 3GYQ A; 1V6Z A; 1P9P A; 1V2X A; 2Z0Y A; 2I6D A; 2QWV A; 3KTY A; 3KNU A; 3QUV A; 3E5Y A; 1VH0 A; 1TO0 A; 2QMM A; 1UAJ A; #chains in the KnotProt database with same CATH homology 3DCM X; 4CNG B; 1X7P A; 1VHK A; 1MXI A; 2O3A A; 2YY8 A; 4E8B A; 2CX8 A; 1NS5 A; 4H3Y A; 4IG6 A; 1X7O A; 3IEF A; 3ILK A; 1UAK A; 3N4K A; 1K3R A; 4FAK A; 1O6D A; 4CNF B; 2HA8 A; 1NXZ A; 1VHY A; 3AXZ A; 1ZJR A; 4CNE B; 3IC6 A; 3NK6 A; 3L8U A; 3ONP A; 3NK7 A; 3AI9 X; 3KY7 A; 1UAL A; 1UAM A; 3AIA A; 1GZ0 A; 3N4J A; 4H3Z A; 1J85 A; 3GYQ A; 1V6Z A; 1P9P A; 1V2X A; 2Z0Y A; 2I6D A; 2QWV A; 3KTY A; 3KNU A; 3QUV A; 3E5Y A; 1VH0 A; 1TO0 A; 2QMM A; 1UAJ A;
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...