
Jmol._Canvas2D (Jmol) "jmolApplet0"[x]
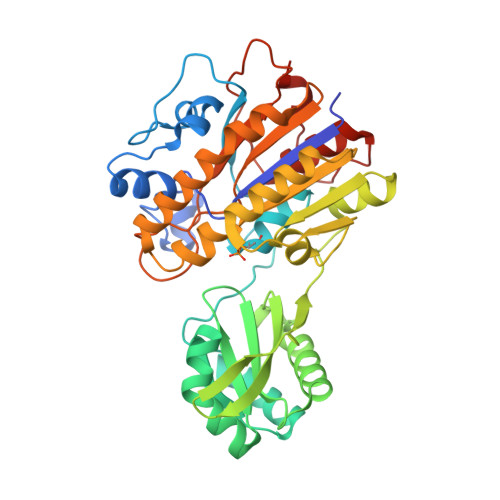
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
+31 | 6-325 | 320 | 1-5 | 342-391 | 326-341 | 5 | 50 | slipknot |
Chain Sequence |
KYKRIFLVVMDSVGIGEAPDAEQFGDLGSDTIGHIAEHMNGLQMPNMVKLGLGNIREMKGISKVEKPLGYYTKMQEKSTGKDTMTGHWEIMGLYIDTPFQVFPEGFPKELLDELEEKTGRKIIGNKPASGTEILDELGQEQMETGSLIVYTSADSVLQIAAHEEVVPLDELYKICKIARELTLDEKYMVGRVIARPFVGEPGNFTRTPNRHDYALKPFGRTVMNELKDSDYDVIAIGKISDIYDGEGVTESLRTKSNMDGMDKLVDTLNMDFTGLSFLNLVDFDALFGHRRDPQGYGEALQEYDARLPEVFAKLKEDDLLLITADHGNDPIHPGTDHTREYVPLLAYSPSMKEGGQELPLRQTFADIGATVAENFGVKMPEYGTSFLNELK
|
Knotoid cutoff: 0.5
Knotoid matrix content: 1
| Knot core range | Knot core length | Knot tails range | Slipknot tails range | Slipknot loops range | N-end length | C-end length | Type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
|
3.1 | 8-329 | 322 | 1-7 | 335-391 | 330-334 | 7 | 57 | slipknot | |
| view details |
|
|
3.1 | 6-334 | 329 | 1-5 | 338-391 | 335-337 | 5 | 54 | slipknot | |
| view details |
|
|
2.1 | 9-327 | 319 | 1-5, 332-391 | 6-8, 328-331 | 5 | 60 | slipknot |
| sequence length |
391
|
| structure length |
391
|
| publication title |
Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
pubmed doi rcsb |
| molecule tags |
Isomerase
|
| molecule keywords |
Phosphopentomutase
|
| source organism |
Bacillus cereus
|
| total genus |
 Genus: 139 Genus: 139
|
| ec nomenclature |
ec
5.4.2.7: Phosphopentomutase. |
| pdb deposition date | 2011-09-22 |
| KnotProt deposition date | 2014-08-18 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF01676 | Metalloenzyme | Metalloenzyme superfamily |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Alpha Beta | 2-Layer Sandwich | Alpha-Beta Plaits | Phosphopentomutase | ||
| Alpha Beta | 3-Layer(aba) Sandwich | Alkaline Phosphatase, subunit A | Alkaline Phosphatase, subunit A |
#chains in the KnotProt database with same CATH superfamily 1KH7 A; 2GLQ A; 3DYC A; 3IGY B; 3T01 A; 1KHN A; 3T00 A; 1SHQ A; 3M8W A; 1AJD A; 1AJB A; 1KH4 A; 1Y7A A; 1AUK A; 1ALK A; 3M7V A; 1B8J A; 1ZEB A; 3BDG B; 3T02 A; 1EW8 A; 3UN2 A; 1Y6V A; 1ELY A; 1E3C P; 3UN5 A; 3TWZ A; 1URB A; 1E1Z P; 1O98 A; 1HJK A; 1ALI A; 1E2S P; 1EQJ A; 2G9Y A; 1KHJ A; 1O99 A; 3IGZ B; 3MK2 A; 1KHL A; 1N2K A; 1P49 A; 1ANI A; 3SZY A; 3M8Y A; 2QZU A; 1ELZ A; 1EJJ A; 3OT9 A; 3UO0 A; 1K7H A; 2GA3 A; 1N2L A; 3UNY A; 1AJC A; 1ZEF A; 2IFY A; 3MK0 A; 1KH9 A; 3BDG A; 1HDH A; 1EW2 A; 3DPC A; 1ALJ A; 1ED8 A; 1EI6 A; 2ANH A; 3UN3 A; 3BDF A; 3ED4 A; 1E33 P; 1EW9 A; 1KHK A; 3SZZ A; 3NVL A; 1ZED A; 1AJA A; 1KH5 A; 3TX0 A; 1FSU A; 3CMR A; 3MK1 A; 1URA A; 3M8Z A; 1ELX A; 1ANJ A; 1HQA A; 1ED9 A; 4FDJ A; 3BDH A; 4FDI A; 1SHN A; #chains in the KnotProt database with same CATH topology 1KH7 A; 2GLQ A; 3DYC A; 3IGY B; 3T01 A; 1KHN A; 3T00 A; 1SHQ A; 3M8W A; 1AJD A; 1AJB A; 1KH4 A; 1Y7A A; 1AUK A; 1ALK A; 3M7V A; 1B8J A; 1ZEB A; 3D7S B; 3BDG B; 3T02 A; 1EW8 A; 3UN2 A; 1Y6V A; 1ELY A; 1E3C P; 3UN5 A; 3TWZ A; 1URB A; 1E1Z P; 3D7S D; 1O98 A; 1HJK A; 1ALI A; 1E2S P; 1EQJ A; 2G9Y A; 1KHJ A; 1O99 A; 3IGZ B; 3MK2 A; 1KHL A; 1N2K A; 1P49 A; 1ANI A; 3SZY A; 3M8Y A; 2QZU A; 1ELZ A; 1EJJ A; 3OT9 A; 3UO0 A; 1K7H A; 2GA3 A; 1N2L A; 3UNY A; 1AJC A; 1ZEF A; 2IFY A; 3MK0 A; 1KH9 A; 3BDG A; 1HDH A; 1EW2 A; 3DPC A; 1ALJ A; 1ED8 A; 1EI6 A; 2ANH A; 3UN3 A; 3BDF A; 3ED4 A; 1E33 P; 1EW9 A; 1KHK A; 3SZZ A; 3NVL A; 1ZED A; 1AJA A; 1KH5 A; 3TX0 A; 1FSU A; 3CMR A; 3MK1 A; 1URA A; 3M8Z A; 1ELX A; 1ANJ A; 1HQA A; 1ED9 A; 4FDJ A; 3BDH A; 4FDI A; 1SHN A; #chains in the KnotProt database with same CATH homology 1KH7 A; 2GLQ A; 3DYC A; 3IGY B; 3T01 A; 1KHN A; 3T00 A; 1SHQ A; 3M8W A; 1AJD A; 1AJB A; 1KH4 A; 1Y7A A; 1AUK A; 1ALK A; 3M7V A; 1B8J A; 1ZEB A; 3BDG B; 3T02 A; 1EW8 A; 3UN2 A; 1Y6V A; 1ELY A; 1E3C P; 3UN5 A; 3TWZ A; 1URB A; 1E1Z P; 1O98 A; 1HJK A; 1ALI A; 1E2S P; 1EQJ A; 2G9Y A; 1KHJ A; 1O99 A; 3IGZ B; 3MK2 A; 1KHL A; 1N2K A; 1P49 A; 1ANI A; 3SZY A; 3M8Y A; 2QZU A; 1ELZ A; 1EJJ A; 3OT9 A; 3UO0 A; 1K7H A; 2GA3 A; 1N2L A; 3UNY A; 1AJC A; 1ZEF A; 2IFY A; 3MK0 A; 1KH9 A; 3BDG A; 1HDH A; 1EW2 A; 3DPC A; 1ALJ A; 1ED8 A; 1EI6 A; 2ANH A; 3UN3 A; 3BDF A; 3ED4 A; 1E33 P; 1EW9 A; 1KHK A; 3SZZ A; 3NVL A; 1ZED A; 1AJA A; 1KH5 A; 3TX0 A; 1FSU A; 3CMR A; 3MK1 A; 1URA A; 3M8Z A; 1ELX A; 1ANJ A; 1HQA A; 1ED9 A; 4FDJ A; 3BDH A; 4FDI A; 1SHN A;
#similar chains in the KnotProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...